11
Pluribiosis and the never-ending microgeohistories
Charlotte Brives
Viruses are defined by scientists as strict parasites, entities which cannot survive without a host organism. Generally speaking, viruses penetrate specific cells within a specific organism and use the metabolic machinery of those cells to power their own reproduction. For some virologists and philosophers of science, the dependency of viruses on their hosts means that they cannot truly be classified as living beings, since they are incapable of surviving alone, they exist ‘on the fringes’ of life. They are not living beings, but nor are they non-organic. The debate is unlikely to be concluded in the near future, since developments in our scientific understanding regularly offer new information which can be difficult to conceptualise. For example, the discovery of the existence of virophages, small viruses that infect larger ones, has put paid to the dogma that a virus can only be a parasite and never a host. It turns out that viruses are also vulnerable to infection.
Nevertheless, it is possible to conceptualise the problem in a different way: a strict parasite cannot be understood without reference to its host, since its very existence depends upon the relationship established between the two of them. In this respect, viruses require us to think beyond the old dichotomy of the living and non-living and instead to embrace a broader understanding of the fundamentally relational nature of biological entities. Although sequencing the DNA of viruses provides insight into their incredible genetic diversity,1 the vast majority of viral characteristics, capacities and competences cannot be experimentally studied, understood or assimilated without reference to the interactions the virus forms with the living species (animal, plant, or bacterial) with which it co-evolves. Their very existence is therefore defined by relationality.
In this chapter, using the specific case of bacteriophage viruses (literally: bacteria-eating viruses) and their bacterial hosts, I explore how taking into account this relational dimension of biological entities can allow us to imagine new therapeutic assemblages. Since their discovery at the beginning of the twentieth century, phages have indeed been used to treat bacterial infections. Although they were neglected in the second half of the twentieth century, notably due to the discovery and then massive production of antibiotics in the 1940s, there has been growing interest in their use since the early 2000s, due to the rise in bacterial resistance to antibiotics.
This revival of phage therapy comes as a counterpoint to the multiple problems posed by the inattentions of production and massive consumption of antibiotics and calls for an examination of the way in which the relationships between humans, microbes and environments are conceived. Whether in bacterial infections or in viral epidemics, as the Covid-19 pandemic has shown and still shows, warlike metaphors predominate (Larson et al. 2005; Brives 2020).
But more generally, the different stories about microbes in social sciences and in biomedicine are often based on opposing duos: war/peace, probiosis/antibiosis, Pasteurian/post-Pasteurian (Paxson 2008, 2011; Lorimer 2017, 2020; for further developments, see the introduction of this book). Even the term amphibiosis – coined by Theodore Rosebury in the 1960s and then used by the microbiologist Martin Blaser to recognise the possibility of a biological entity being friend or foe, depending on the context – does not extend beyond this binary view of relations between species (Blaser 2014). This is because these narratives are based on a relatively fixed conception of biological entities, and thus make it impossible to think about how their relationships, in one way or another, transform them.
Thus, the recognition of the variability of relations between humans and microorganisms, as this collective volume testifies, may be an important step but it is also a question of going beyond a fixist conception of both relations between species (by recognising that the relational status of ‘friends’ or ‘enemies’ is spatially and temporally located) and of the species themselves (by taking into account the transformative potential of interspecific encounters).
In this chapter, I describe the practices of isolation and collection of new bacteriophage viruses, which are essential to the development of phage therapy. In particular, I show how the collection constitutes the capture at a particular point in time, for reasons specific to the experimenter (and by extension to the functioning of phage therapy), of a constantly evolving relationship between a virus, a bacterium and a human. If not conducted carefully, however, this capture can lead to an essentialisation of the entities thus collected. It is then necessary to integrate this work of collection into a wider account of the relationships between humans, viruses and bacteria.
Observing and learning from viruses and bacteria gives us an opportunity to understand the term ‘pluribiosis’. Pluribiosis is the recognition of the existence of multiple relational spectra between entities forever in the process of becoming, constantly shaped and transformed by their interactions with other living things, and by the context in which they occur.
In what way can this form of attention to the relations that pluribiosis represents help us develop alternative conceptions of health and ways of treating infections? Because it involves at least humans, viruses, bacteria and environments – according to temporalities specific to each biological entity – phage therapy offers us narratives that refuse fixity and recognise the situated knowledge (Haraway 1988) and situated biologies (Niewöhner and Lock 2018), and therefore the necessarily situated character, of their applications in biomedicine.
This chapter is informed by three years of fieldwork with agents in phage therapy (researchers, clinicians, patients, regulatory agencies) in France, Belgium, and Switzerland, as well as by my membership of bacteriophage virus research networks.
How to play with the potentialities of phages

Fig. 11.1 Lytic and lysogenic cycles of bacteriophage viruses (credit: Tristan Ferry, Hospices Civils de Lyon, Phages In Lyon)
Phages and bacteria have been co-evolving in dynamic and complex ways from their origins. Phages are the most numerous forms of biological entity on earth. It is estimated that at any given time, 40% of bacteria on the planet are infected with a virus. However, the relationships between phages and bacteria are highly specific: a virus is generally only capable of infecting a single species of bacteria. Some are even specific to just one of the genetic variants of that species. Moreover, when a given bacteriophage comes into contact with a population of clonal bacteria (genetically identical bacteria), the consequences are not always the same (see Figure 11.1). Some of the bacteria will die, destroyed by the virus which has used them in order to self-multiply (lytic cycle). Others will learn to live with the virus, having acquired the ability to withstand infection either through mutation or selection. Others still will develop a profoundly intimate relationship with the virus: the latter will become integrated into the genetic material of its host, either temporarily or permanently (lysogenic cycle). If they do finally part ways, the virus may leave behind some of its genetic material and/or take away part of the bacteria’s genome, leaving both entities transformed as a result (this is notably how viruses participate in the spread of antibiotic resistance genes in bacteria through transduction phenomena).
The bactericidal potential of phages has been used in therapy since their discovery in 1917 (d’Hérelle 2017). However, it was not until the second half of the twentieth century that the resources offered by phage therapy were understood.2 Today, a consensus exists around the use of phages for therapeutic purposes, based on the use of strict ‘virulent’ phages (phages that perform lytic cycles and are unable to enter a lysogenic one when they encounter the bacteria of interest). Temperate phages could indeed give the bacterium new skills, including the impossibility of establishing any other relationship whatsoever with an identical phage.
The principle of the therapy is simple: isolate the bacterial strain responsible for the infection in a human, then find a strictly virulent phage active on this bacterium and administer it to the patient. Since phage therapy is still experimental, the process is generally split: hospital infectious diseases specialists isolate the bacterial strain, then send it to one or more research laboratories to determine whether they have phages active against the pathogen in their collections. Then, once one or more active phages have been isolated, characterised and produced according to standards ensuring their biological quality, they are sent to the hospital to treat the patient. This chapter is devoted to the isolation and collection of new phages. The availability of such collections is a sine qua non condition for the development of phage therapy. A collection may place a focus on the scientific and technical processes involved, but it also constitutes an ontological bifurcation, which can lead to different models of development.
The ethnographic part of this chapter is based on fieldwork carried out in a Swiss laboratory in November 2019. The team was led by Jim, a biologist who has been working on phages for over 20 years.3 For several years now, Jim’s team has been working on the creation of collections of virulent phages active on multidrug-resistant (MDR) bacterial species considered problematic in terms of public health by the WHO.4 Jim almost never works in the lab. Most of his time is now spent filling out funding requests. I therefore spent most of my time with Julie, the team’s technician, who does almost all the lab work, observing her manipulating phages and bacteria.
Shortly before I arrived in the laboratory, Jim received two strains of two different bacterial species responsible for the infection of a patient being treated at a university hospital in France. The patient was infected with a strain of Klebsiella pneumoniae (KP) as well as a strain of Proteus miserabilis (PM), both antibiotic-resistant. Faced with the failure of successive treatments, the doctors in charge of the patient turned to Jim to try phages. However, the laboratory did not have any phages active on Proteus, a bacterium with which it does not usually work, and the phages they had for Klebsiella were ineffective on the patient’s strain. Jim and Julie decided to find phages active against these two bacteria to send to the university hospital.
There are different techniques for working with phages and bacteria. During my presence in the laboratory, Julie worked mainly in a solid environment, i.e. with petri dishes. In general, the techniques are relatively simple: Julie first pours a culture medium into large square petri dishes, to which she adds a bacterium of interest. The culture medium slowly hardens and forms a transparent agar. Normally, if this dish is then placed in an incubator at 37°C, the bacteria will reproduce. Thus, after a few hours, a bacterial mat can be observed. The agar then has an opaque appearance, proof of the presence of bacteria evenly distributed over the petri dish (see Figure 11.2). If, before placing this dish in the chamber, drops of solution containing phages are placed on the hardened agar, then we can observe plaque forming units (PFU): small transparent holes in the bacterial mat (see Figure 11.3). There are no more bacteria in these areas. They have been lysed.

Fig. 11.2 Petri dish containing a nutrient medium and a bacterial strain. After 24 hours in an incubator at 37°C, a uniform opacity can be observed due to the development of the strain, which forms a mat (photograph by Charlotte Brives, 2019).
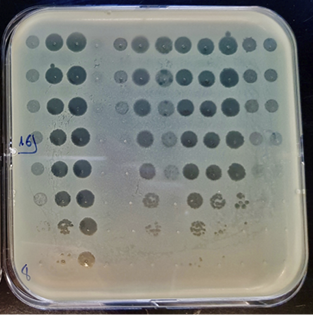
Fig. 11.3 Petri dish on which drops containing bacteriophage viruses have been placed before placement in the incubator. We can observe plaque- forming units (PFU): the bacteria have been destroyed, their absence indicating the presence and activity of the bacteriophage viruses (photograph by Charlotte Brives, 2019).
Julie uses these techniques, which seem simple at first glance, to isolate new phages. But where to find them? As mentioned, phages, being strict parasites of bacteria, are found everywhere that bacteria live. The best place to find them is in so-called rich waters – sewage or treatment plants, for example. Over the past three years, I have learnt that not all waters are equal. A technician working in a start-up told me water withdrawals downstream of the chemical industries should be avoided. And Julie mentioned: ‘hospital waters work well, much better than anything else. We’re very happy only looking in hospital waters’. Hospitals are overflowing with bacteria, which are most often multidrug-resistant due to the high selection pressure exerted by repeated antibiotic treatments on patients.
To this must be added the relational dimension of phages, which Julie repeated several times during my fieldwork: a phage cannot be thought of alone. It is always a phage/bacteria pair. So, when I asked her if, because of the co-evolution between phages and bacteria, it was not more interesting to look for phages in waters near where the patient fell ill, Julie gave this answer:
Yes, most of the time that’s an easier option. Because the population around you potentially has the same bacteria. We know that generally speaking the bacteria we find in Europe will not be the same as in the United States, so the waters will not work in the same way and there is little chance that a phage from the United States will work as well on our collections.
This territorialisation of the relationships between phages and bacteria was exemplified by Julie in the case of a patient hospitalised in a city close to where Jim’s team is located. Several laboratories in Europe were asked to find an active phage against the bacteria responsible for its infection. Jim’s lab, near the city where the patient was hospitalised, was the only one with phages that showed positive results on the patient’s bacteria. This example is often brought up by various actors I have met in recent years to highlight the ecosystemic dimension of phage therapy.
We are therefore witnessing a geographical discrimination of waters, which reflects the co-evolutionary relationship of these two biological entities; the best place to find an active phage on a pathogenic bacterium is near the origin of this pathogenic bacterium.
However, Julie went even further, mentioning that she had often observed that French waters generally work better than Swiss waters, and she had developed a hypothesis for this:
In France, we find them more easily than in Switzerland so I wonder if they use certain water treatment processes. Or perhaps the Swiss are cleaner than the French, perhaps they have more hygiene regulations than in France. I did wonder that. I think they may be cleaner than the French.
For Julie, not all waters are equivalent, and the factors involved are very diverse. The therapies used, hygiene rules, public health policies and water treatment may influence the presence and therefore existence (or not) of phages and their diversity.
It was by pouring rich waters onto the dishes and making repeat observations that Julie developed explanations and arguments for the richness of waters and the continuum between the micro- and macro-biology of entities and multispecific communities.
She poured a nutrient medium into several petri dishes, to which she also added her bacteria of interest, either KP or PM. She then poured water from different sources onto each of the dishes and placed them in an incubator at 37°C for 24 hours. The next day, she looked carefully at her dishes for PFU. Wherever there was a hole in the mat, and therefore probably a phage, Julie took a sample. She prepared a test tube in which she put a liquid nutrient medium, the bacteria of interest and the sample taken on the PFU. She then placed everything in the incubator for three hours during which the bacteria (and perhaps the phage) reproduced. Finally, she centrifuged the tube for ten minutes.
The heavy bacteria were then concentrated in a pellet, a small deposit at the bottom of the tube. She recovered the supernatant, which she filtered to remove bacterial debris. Part of this filtrate was placed back on a petri dish containing the nutrient medium and the bacteria of interest. If there were still PFU after another round of incubation, she would repeat the process: sampling, incubation, centrifugation, filtration. Julie carries out between three to eight cycles (sampling, incubation, centrifugation, filtration) in order to isolate a phage. According to her, performing these separate cycles means she can be sure that the sample contains just one phage and that it is active.5
During these different cycles, Julie learnt a lot about her phages and bacteria: the ideal moment to put them in contact, the length of this contact time and the optimal temperature. The methods used may have been standardised, but she learnt to take into account not simply the characteristics of a phage and a bacterium but also the characteristics of their encounter – another reminder that each encounter, each relationship, is unique.
Julie summed this up during our discussion in the lab:
You have to grope about! You grope about and you have to test different methods. You might play with the temperature a little bit. It’s a lot of trial and error. Everyone says it’s super easy [to produce phages], and overall, it is easy. But you might have to make small adjustments. If you want phages that all have certain characteristics, you’ve got to work with the phage.
‘Work with the phage’. Each phage/bacteria pair is unique, and so is the relationship between the technician and a given pair. The skills and know-how Julie has acquired from being in contact with microorganisms are essential. Some bacteria, such as Staphyloccocus aureus, are described by the technician as ‘capricious’, others as ‘permissive’. Some do not ‘work properly’, which can mean that ‘your phages will not perform as well’. Thus, it is a question of taking into account the particularities of each relationship and the way in which historicity is embodied in the biology of each organism.
Naming a phage, or the snapshots of microgeohistories
After these various stages, Julie obtained three active phages against the patient’s KP strain and two phages for the PM strain.
These phages would later be sequenced to know their genome, but she was already convinced that the three anti-Klebsiella phages were different. In addition to having been isolated in waters of different origins, she showed me the PFU: ‘they don’t have the same morphology at all. Typically, for KP95, I’ll have three different phages. These are good ones. This one’s very tiny, it’s not very clear’.
What she could see with the naked eye, with disconcerting ease, required my close attention, even though she had told me what to look for: the size of the PFU, the regularity of their edges, their opacity. Julie had developed a level of intimacy with the lab’s phages that even Jim envied.
Her statement, ‘They do not have the same morphology at all’, is simplistic because it is in fact the PFU, the holes in the bacterial mat, and not the phages, which do not have the same morphology. The phages cannot be observed with the naked eye. This is yet another reminder that what was being evaluated was in fact a relationship, not a biological entity as such, whose presence and skills were only visible through their effects on bacteria and deeply mediated by the technician’s perspective.
This point is illustrated by the assignment of a name to each of the new phages Julie had isolated. One of the anti-Klebsiella phages was named 4035-KP95. All the phages in the laboratory starting with 4000 are Klebsiella phages. It was therefore the 35th Klebsiella phage in the collection. The segment ‘KP95’ corresponds to the name of the bacterial strain from which the phage has been isolated. This reinforces, if necessary, the strong relational dimension and the historicity of each entity. Since phages and bacteria co-evolve permanently, if this phage were to come into contact with another bacterium, say KP112, the resulting phage could be given the name 4035-KP112, to take into account its probable evolution. It would no longer be exactly the same phage, as Julie explains: ‘it’s a phage-bacterium pair. Change your bacteria or anything else, and your results will be different. Your phage will not react in the same way’.
This leads us to another conclusion concerning the phage’s entry into a collection: the act of naming the phage is not only a scientific ‘birth’ act of the phage, but above all an act of fixation, crystallisation, essentialisation of a relationship. The technician decides, at a given time, after bringing phages and bacteria together, to put an end to a relationship that is destined to constantly evolve. At that precise moment, the technician considers that the phage she has in front of her perfectly expresses the potential she wanted to exploit in its relationship with the bacteria of interest: this phage is virulent, exhausting (almost) all the bacteria it encounters in order to reproduce. This phage and this bacterium will then join the laboratory’s collection. Each will be placed in tubes and stored in two freezers, to which only the technician and the team leader have access, at -80°C.
Once in the collection, all this work of isolation and characterisation must not be in vain: nothing must change. Patients’ bacteria, like the phages of sewage water, must remain alive but no longer evolve, whether on paper or in a test tube because what the technician knows is only valid for this particular pair, at this particular moment in their relationship.
During the experiments, everything is based on relationships and everything happens because there are co-evolutionary relationships, therefore entry into the collection sanctions the objectification of microorganisms. The assignment of a name implies a marked identity. Each new phage is then sequenced, and the sequence of nucleic acids sets this identity in stone.
In this context, the laboratory’s collections can be seen as snapshots of microgeohistories. Each phage has a history: it is isolated from a water sample collected in a specific place and at a given time, on the bacterial strain of a patient who themselves has a complex history, sometimes in a nearby area, sometimes not. Other phages, such as the hypothetical 4035-KP112, will result from the encounter between one of the phages in the collection and a new bacterium, perhaps taken from a person suffering greatly from its presence in his or her body. Collections are therefore the fixation, at a given moment, of a tripartite human/bacterial/phage relationship.
The colossal level of work required to maintain these collections, and the precautions surrounding their handling, however, testify to their scientific and therapeutic value as well as their precarity. During a laboratory ethnography conducted on the relationship between humans and Saccharomyces cerevisiae yeasts, I showed how access to the -80°C freezer, which contained the laboratory’s yeast collection, was strictly regulated. The comings and goings around the freezer, the organisation of the laboratory, its implicit and explicit norms, the evident rules of laboratory life ensured that what was in the freezer, the yeast strains, remained identical to themselves and could continue to constitute ‘reliable witnesses’ to the experiment, to use Isabelle Stengers’ expression (Stengers 1993; Brives 2017). In the Swiss laboratory I worked in, it was much the same. Everything is rendered more complicated because of the particularities of phages and bacteria, and the relational nature of the collections. When a particular phage has been isolated from a particular bacterium, both must be preserved.
These collections are precious. They are set up in order to create the greatest possible diversity of phages available in order to be able to treat patients. But it is important to remain cautious and to take them for what they are: snapshots of never-ending microgeohistory, never-ending multispecific dances. And yet phages are still most often presented, in conferences or writings aimed at the general public, as ‘professional killers’ of bacteria, or as ‘snipers’. These metaphors are far from insignificant and can, as we shall see, guide the development of phage therapy in one direction or another.
Pluribiosis, or the creative powers of the living
The relational capacities of phages and bacteria have been observed and utilised in laboratories for decades. They have powered the development of molecular biology (Kay 1993; Morange 1994), opening up vast new horizons for both fundamental and applied research (for example the ability to produce genetically modified organisms, therapeutic molecules, biofuels, etc.), as illustrated by the award of the 2020 Nobel Prize in Chemistry to Emmanuelle Charpentier and Jennifer Doudna for their work on the CRISPR-Cas9 system. Many scientists do not consider phages as life forms, but simply as tools in the service of human needs. The manipulation of ‘life’ as a soulless, disembodied force has been the stuff of countless science fiction stories and dystopian visions, more often than not used to castigate mankind’s tendency to objectify and commodify nature, a force which remains exasperatingly external to humanity. For other thinkers, these capabilities are a constant source of new questions regarding the spectrum of life, cooperation, evolution and ecology. Studying viruses and bacteria reveals far more than just a spectrum of relational mechanisms. It lays bare the essential plasticity of these entities, their shared existence in and through the relationships they form. Thanks to the fusion of its genetic material during a lysogenic cycle, the mode of existence of the virus changes radically. It becomes an integral part of the host bacterium. Some papers sometimes describe these viruses as being ‘dormant’, which could give the impression that during this time the virus is not doing anything. Meanwhile, the bacterium in which the virus is ‘dormant’ develops new capacities. To whom do they belong? The bacterium or the virus? Do the two entities still exist independently of one another, or have they become a new entity entirely, a hybrid? These questions are rendered even more complex by the fact that the new entity thus formed may enjoy only a fleeting existence. In certain conditions, the viral genetic material may separate from that of the bacterium, replicate itself hundreds of times to form new viral particles and break free, killing the bacterium in the process. Can the resulting entities be considered the same virus, or the same bacterium?
Once again, metaphors play an important role here. It is difficult to express ‘viro-bacterial’ behaviour in words without having recourse to the familiar vocabulary of sexual reproduction and immunology – both highly gendered discourses, as anthropologist Emily Martin has demonstrated (Martin 1991, 1995). Scientists talk of phages ‘injecting’ their DNA, ‘penetrating’ the host cell, ‘lurking’ or ‘concealing themselves’. They describe them as ‘sleeper agents’ in bacterial DNA, when they are not acting as ‘professional killers’ or ‘snipers’. The vocabulary used is all about transgression, violation, insidious conflict and destruction, while phage behaviours could be framed very differently, sometimes by the same researchers, talking about their incredible potentialities and the way they engage in kinky sex, ‘homologous and illegitimate recombination with related and completely alien genomes, orgies of hundreds of genes’ (Rohwer et al. 2014). Some virologists, having become intimately familiar with the world of phages and bacteria, are reluctant to say that the phage even ‘kills’ its host. In many cases, the bacteria simply die because all of its resources have been used up by the virus. These reformulations reveal the possibility of an entirely different narrative: one in which the virus does not intentionally kill the bacterium but instead uses it as a matrix for a process of replication and creation which incorporates fragments of the bacterium itself. A process of creative forces breaking free of the dogmas of sexual reproduction and immunology, unfettered from notions of self and other, organic intimacies which reveal the porous nature of the categories upon which modern Western thought relies to understand and control the world. This hints at the possibility of a more subtle spectrum of relations between entities, which are increasingly difficult to essentialise. And in a further layer of complexity, we must bear in mind that these relationships also depend upon the broader milieus within which they exist, and which they help to shape and compose, sometimes with spectacular results.6
These modes of interaction are not specific to viruses and bacteria. Between 5% and 8% of the human genome is of viral origin, i.e. created by the fusion, at one time or another in the evolutionary history of this species, of the genetic material of a virus with that of an ‘infected’ host cell. The most well-known example in the animal kingdom is the existence of syncytins, proteins essential to placental development. This capacity is specific to mammals but was in fact – and this is only a paradox if one is committed at all costs to maintaining the distinction between the ‘essence’ of a living thing and its pollution by external elements – made possible by interactions with a virus in the distant past (Dupressoir et al. 2012). This DNA of viral origin, much of which is considered to be ’junk DNA’ because of its apparent (read immediate, current) uselessness, can be seen as a sort of fossil record of relational experiences – a precious archive of the events which led mammals to their current state.
The essentialisation of entities and relations is only possible if you stop their movement or slow it down sufficiently (when you do not simply freeze it as we saw in the lab). In this respect the microbial world is precious. Without claiming to account for all of the differences which exist between these forms of life, we can at least point out that the time frames in which they operate are clearly distinct from our own. The span of one human life is time enough for thousands of generations of bacteria, billions of cells living and interacting with bacteriophages, engaging in mutual transformations and acting upon the milieus which they inhabit. In short, bacteria work much faster than humans do and, in doing so, they allow scientists to observe processes which require painstaking reconstruction and considerable guesswork in the animal and plant kingdoms.
The importance of movement and the realisation that it is impossible to think effectively without it is fundamental to understanding viruses and bacteria. It is only the distortion effect induced by the slowing down of time that causes us to imagine univocal relationships between entities whose essential identities remain fixed, imagining the milieus in which they exist (and which they in fact help to shape and transform) to be simple backdrops or static scenery, when they actively participate in their transformation.
The observation of viruses and bacteria does not lead to an essentialisation of entities, relationships and milieus; rather, it allows us to consider pluribiosis: the entanglement of multiple situated relational spectrums that involve entities and milieus that are always in the process of becoming, constantly formed or transformed by interactions.
If essentialisations are necessary for scientific activity (and, more generally, action), the notion of pluribiosis reminds us that they are situated and fixed instantiations, snapshots, of the fundamentally relational nature of life. Reporting/focusing/accounting for situations and relationships that the different actors – biologists, ecologists, physicians, regulators, patients – that I have met or read about describe and go through, then, makes it possible to grasp the diversity and becomings of entities.
Pluribiosis is a heuristic concept because it recognises the importance of relations and milieus and, thus, prevents us from assuming what is biological or social, natural or cultural. It maintains a close attention to what the entities become and the transformations that shape the situations described and what they contain. It is a prescriptive concept as well, insofar as it recognises the fundamentally multiple and situated nature of knowledge about life (pluri-biosis) but also the transformation potential of these knowledges. If evolution and relation are constitutive of life, then the knowledge that claims to account for it, as well as its uses, must be included in the configurations in which it is produced. As the ethnography of our collection of bacteriophages has shown, this is an element that should not be overlooked in the development of phage therapy.
Towards a pluribiotic medicine
Paying attention to the forms of interaction between phages and bacteria, and observing laboratory practices that allow the isolation and selection of phages for their capacity to develop one form of interaction to the detriment of others, encourages thoughts of another conception of medicine and care. This is a medicine that I have chosen to describe as pluribiotic, considerate of the never-ending histories of multispecific assemblages which are always in the process of becoming.
Because they never lose sight of the evolutionary and creative capacities of the living, researchers and physicians condition therapeutic success on the use of not one but several phages for a given bacterium. Although a phage can be selected and ‘trained’ for its lytic abilities, the creative powers of living things cannot be controlled and mastered. Once brought into contact at the time of patient treatment, phages and bacteria will continue to explore a spectrum of relationships that can never be completely reduced to what humans want to do with them. The disappearance of the pathogenic bacterium can be ensured by the joint action of a single phage and the human immune system. The former will drastically reduce the bacterial population, the latter will then be able to effectively treat the infection. Although different phages chosen by humans because of their virulence on the pathogenic bacteria are administered, each one will start its own dance with the latter, each one will use it as a fertile matrix to reproduce, reinvent itself, metamorphose. The bacterium will then no longer be able to interact with the phages long enough to find new assemblages, new becoming-with (Haraway 2003). It will disappear.
For these reasons, the availability and the success of phage therapy depends on the constitution of phage collections. The practices that enable these collections are therefore fundamental and at the heart of the development of this alternative/complement to antibiotics, as are the issues they raise. For those who are not attentive, for those who do not grasp the resources offered by pluribiosis and the patient adjustments made between phages, bacteria, humans and the environment, collections can appear as a manna: reified, controllable, standardisable, exploitable and mass usable entities. If neglected, collecting appears to be an end, not a necessary detour. The microgeohistories at work are then frozen in History, that of Human control over Nature.7 The collecting of phages becomes a tipping point, a possible bifurcation between radically different projects and conceptions of infection, care and the production of new therapies.
This is precisely where the possibilities of phage therapy come into play: in the choices we make in the way we approach and use these collections.
Currently, there is some form of consensus on the possibility of developing two models of therapy, which can easily be linked to a differential use of collections and to opposing conceptions of the living: the ‘sur-mesure’ and the ‘prêt-à-porter’ models (Pirnay et al. 2011). The first involves the selection, within collections, of highly virulent phages on the bacterial strain responsible for the infection. It may also involve adjusting these phages to the bacterial strain, by ‘training’ the phages to increase their virulence. In this model, the assemblages between phages, pathogenic bacteria and humans come first. The second model decontextualises the infection. Favoured in particular by start-ups, it is based on the development of cocktails containing a few phages that show activity on a wide variety of strains of a given bacterial species. The collections can help to identify such phages. These phages could then be produced, marketed and administered en masse. In this ‘prêt-à-porter’ conception of treatment, which can also be described as a ‘one-size-fits-all’ approach, the living, co-evolutive dimension is unthinkable. The objectified phage becomes an umpteenth antibiotic and is used as a chemical molecule (Brives and Pourraz 2020). The phages of this type of cocktail could then be only partially active and, for example, infer the selection of resistant bacteria.
The sometimes deregulated and often unquestioned use of antibiotics in human and animal health has, however, largely led to the rise of antimicrobial resistance (AMR). What has been forgotten is precisely the dynamic, relational and adaptive nature of living things, the creative power of the living. Science historian Hannah Landecker (2016: 21) has shown how the antibiotics industry has completely changed the biology of bacteria:
The bacteria of today are not the bacteria of yesterday, whether that change is registered culturally, genetically, physiologically, ecologically or medically. Bacteria today have different plasmids and traits and interrelations and capacities and distributions and temporalities than bacteria before modern antibiotics. It is not even clear that ‘bacteria’ remains the only or the most salient category with which to think about antibiotic resistance. This biological matter, chewing away its own ontology, is historically and culturally – and materially – specific to late industrialism, produced in and by previous modes of knowledge.
AMR, to which phages participate when developing lysogenic relations to bacteria, can be seen as the manifestation of pluribiosis, of the creative power of the living. These creative powers are not in themselves good or bad. They simply create new assemblages. The living acts and reacts. Faced with chemical molecules, bacteria have evolved very quickly towards new and cumulative forms of resistance. What if, instead of chemical molecules or in addition to chemical molecules, bacteria were massively exposed to standardised phage cocktails, biological entities that also have tremendous evolutionary capacities?
The deeply relational nature of living things is a forgotten element in antibiotic therapy. Phages, by their particularities, help us to remember this dimension and to develop, as many agents in phage therapy hope, a medicine that actively takes pluribiosis into account.
It is therefore a matter of making a choice to be attentive to pluribiosis; to contextualise within never-ending microgeohistories; and to renounce both ‘one-size-fits-all’ solutions, which will imply creating new models of development (Brives and Pourraz 2020). Also to be renounced is the perpetuation of the story of Human control over Nature, the possible consequences of which humans can no longer ignore.
Notes
1 See the phylogenetic map available at: http://virusmap.univ-lyon1.fr/.
2 For more details on a historical approach to bacteriophages and phage therapy, see the special issue edited by Neeraja Sankaran (2020).
3 For reasons of confidentiality, Jim’s identity, his laboratory and the city in which it is located have been removed.
4 These bacterial species are known under the acronym ESKAPE, for Escherichia coli, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter sp.
5 Confirmation of the purity of the phage will come with sequencing and a more complete characterisation of the isolated phage, a stage in the process that I do not cover in this article.
6 For an example of the involvement of bacteriophage viruses in trophic chains, see Peduzzi et al. (2014).
7 In Facing Gaïa, Eight lectures on the new climatic regime (2017), Bruno Latour uses the term gaïahistoire, which he contrasts with History. Donna Haraway takes up this distinction between geohistory and the History of Man’s control of Nature in Staying with the Trouble (2017).
Acknowledgements
I thank the Kilpisjärvi Collective, Alexis Zimmer and Bruno Latour for comments on earlier drafts of this text. I acknowledge the support of the Agence Nationale de la Recherche (grant no ANR-18-CE36-0001) and the Région Nouvelle-Aquitaine (grant no 2018-1R40218).
References
Blaser, M., Missing Microbes (New York: Picador, 2014).
Brives, C., ‘Que font les scientifiques lorsqu’ils ne sont pas naturalistes ? Le cas des levuristes’, L’Homme, 222 (2017): 35–56.
_______ ‘The Politics of Amphibiosis: The War against Viruses Will Not Take Place’, Somatosphere (2020) <http://somatosphere.net/2020/the-politics-of-amphibiosis.html/> [accessed 6 November 2020].
Brives, C., and J. Pourraz, ‘Phage Therapy as a Potential Solution in the Fight against AMR: Obstacles and Possible Futures’, Palgrave Communications, 6.100 (2020).
Dupressoir, A., C. Lavialle, and T. Heidmann, ‘From Ancestral Infectious Retroviruses to Bona Fide Cellular Genes: Role of the Captured Syncytins in Placentation’, Placenta 33.9 (2012), 663–71.
Haraway, D., ‘Situated Knowledges: The Science Question in Feminism and the Privilege of Partial Perspective’, Feminist Studies, 14.3 (1988), 575–99.
_______ The Companion Species Manifesto: Dogs, People, and Significant Otherness (Chicago: Prickly Paradigm Press, 2003).
_______ Staying with the Trouble, Making Kin in the Chthulucene (Durham, NC: Duke University Press, 2016).
d’Hérelle, F., ‘Sur un microbe invisible antagoniste des bacilles dysentériques. Compte-rendu de l’Académie des Sciences’ 165.11 (1917), 373–5.
Kay, L., The Molecular Vision of Life (Oxford: Oxford University Press, 1993).
Landecker, H., ‘Antibiotic Resistance and the Biology of History’, Body & Society, 22.4 (2016), 19–52.
Larson, B. M. H., B. Nerlich, and P. Wallis, ‘Metaphors and Biorisks: The War on Infectious Diseases and Invasive Species’, Science Communication, 26.3 (2005), 243–68.
Latour, B., Facing Gaïa: Eight lectures on the New Climatic Regime (Polity Press, 2017).
Lorimer, J., ‘Probiotic Environmentalities: Rewilding with Wolves and Worms’, Theory, Culture & Society, 34 (2017), 27–48.
_______ The Probiotic Planet: Using Life to Manage Life (Minneapolis: University of Minnesota Press, 2020).
Martin, E., ‘The Egg and the Sperm: How Science Has Constructed a Romance Based on Stereotypical Male-Female Roles’, Journal of Women in Culture and Society, 16.3 (1991), 485–501.
_______ Flexible Bodies (Boston: Beacon Press 1995).
Morange, M., Histoire de la biologie moléculaire (Paris: La Découverte, 1994).
Niewöhner, J., and M. Lock, ‘Situating Local Biologies: Anthropological Perspectives on Environment/Human Entanglements’, Biosocieties, 13 (2018), 681–97.
Paxson, H., ‘Post-Pasteurian Cultures: The Microbiopolitics of Raw-Milk Cheese in the United States’, Cultural Anthropology, 23 (2008), 15–47.
_______ The Life of Cheese: Crafting Food and Value in America (Berkeley: University of California Press, 2012).
Peduzzi, P., and others, ‘The Virus’s Tooth: Cyanophages Affect an African Flamingo Population in a Bottom-Up Cascade’, ISME, 8.6 (2014), 1346–51.
Pirnay, J. P., and others, ‘The Phage Therapy Paradigm: prêt-à-porter ou sur-mesure?’, Pharmaceutical Research, 28 (2011), 934–7.
Rohwer, F., and others, Life in Our Phage Wordl: A Centennial Field Guide to the Earth’s Most Diverse Inhabitants (San Diego: Wholon, 2014).
Sankaran, N., ed., Diversifying the Historiography of Bacteriophages, Notes and Records: Special Issue, 74.4 (London: The Royal Society, 2020).