8
Ontologies of resistance: Bacteria surveillance and the co-production of antimicrobial resistance
Nicolas Fortané
The overuse and misuse of antibiotics in human medicine and in agriculture are key factors behind the development of resistant bacteria that generate infections which are increasingly difficult or even impossible to cure (O’Neill 2016). In the farming sector, although some half-hearted measures were introduced in the 1970s, it was only at the end of the twentieth century that ambitious regulations began to be implemented in order to better control the use of antibiotics in livestock, particularly the 2006 European ban on antibiotics as growth promoters (Kirchhelle 2020).
These public policies are also important for having introduced a series of lesser-known measures, namely surveillance systems for monitoring resistant bacteria in livestock farms and slaughterhouses, along with other devices for monitoring the sale of antibiotics in veterinary medicine. Most European countries have created such surveillance and monitoring systems, which are regularly updated to more effectively track the evolution of antimicrobial resistance (AMR) and to provide a basis for public policy in this area. However, many outstanding issues related to the functioning of these systems remain. They range from the type of data produced and the methodologies used by these systems to how AMR is defined, and the kind of human-microbe relationships involved.
In recent years, numerous studies have explored the history of epidemiological surveillance. They have shown how the globalisation of these surveillance systems is linked to the profound changes in risk management and assessment policies that took place at the turn of the twenty-first century. From a biosecurity standpoint (Lakoff and Collier 2008), the deployment of surveillance networks was linked to the emergence of new actors, knowledge and techniques that helped to reframe numerous public health issues. Although the roots of this dynamic could be traced back to the post-World War II period and the development of epidemic intelligence (Fearnley 2010), the logic of preparedness that was imposed from the late twentieth century onwards considerably reshaped global health policies and the way surveillance systems operate (Lakoff 2010). While this chapter draws from these studies, it also aims to give them a fresh perspective.
The rise of epidemiological surveillance in response to biosecurity concerns was less a rupture than an adjustment of existing systems, particularly with regard to risks involving animals. Major institutional reforms involving both national and international governance of public health certainly took place in the late 1990s. However, what I would like to highlight here is that there was no massive shift or change in the design of policy instruments. Instead, there was a gradual rearrangement of pre-existing infrastructures and a reorientation of the activities taking place within them. Biosecurity concerns relating to animal surveillance were tacked on to systems initially created for other purposes, such as academic research, the technical management of agricultural or veterinary practices, or the conservation of biodiversity (Fortané and Keck 2015). Surveillance thus relates not only to practices of control but also to practices of care (Manceron 2013).
Second, and consequently, to understand how different activities coordinate or compete within surveillance systems, we also need to understand them ‘from the bottom up’, i.e. through the everyday practices that help to structure surveillance systems and bind together all of the actors (humans and non-humans) involved in their operation. In short, we must examine not only the knowledge and policy instruments that built these surveillance systems but also the practical activities and the ‘disciplinary borderlands’ (Enticott 2017) that ensure the ordinary fabric of the phenomenon that they are meant to monitor and govern (Fortané and Keck 2015). In this chapter, I therefore intend to demonstrate how the surveillance of AMR relies on an assemblage of various forms of practices related to animal health, from animal care performed by vets and farmers to biological research and epidemiological surveys performed by scientists.
To examine the material conditions behind the production of monitored and governed objects more closely, we may look at certain recent works which extend this pragmatic perspective via the ‘ontological turn’ in the anthropology of science and/or medicine (Woolgar and Lezaun 2013). These works highlight the different stages in the construction of an object of knowledge, such as the wolf population (Doré 2013) or foot-and-mouth disease (Law and Mol 2011), which, through these successive operations, also becomes an object of policy, i.e. a governable object (although this governability may in part remain uncertain). To consider these different approaches, I use the notion of apparatus to refer to the surveillance system studied here. In effect, such an apparatus works as an ‘interessement device’ (Callon 1986) which allows the assemblage of different forms of practice and therefore engagement; both humans (veterinarians, microbiologists, and epidemiologists) and non-humans (animals, microbes, and antibiotics), contribute to the production of AMR. AMR thus acts as a ‘boundary-object’ (Star and Griesemer 1989) that is able to bring together the multiple actors, tools and modes of knowing which are enacting the apparatus and providing the material conditions of existence of this boundary-object. These enactments through which the surveillance system exists, and AMR is somehow brought to life, are precisely what this chapter aims to analyse, as they refer to the relationships that bind together humans, animals and microbes.
However, as said above, the practices and forms of engagement through which the apparatus is enacted can be quite various. Veterinarians, biologists and epidemiologists do not have the same relationships with animals and microbes. The surveillance system can thus produce different versions of AMR, or what I will call here ‘ontological referents’ (Doré 2013). Even more importantly, the surveillance system can only work and reach its final objectives because several ontological referents are being produced, as a consequence of the variety of practices and engagements that are assembled in the diverse activities that make the system work (data collection, analysis, etc.). In the case that is presented here, I will show that the population of resistant bacteria that epidemiologists are trying to measure and construct as a ‘governable’ object is not the only form of AMR that is enacted. Veterinarians and microbiologists, whose engagement is likewise vital to the operation of the system, also produce their ‘own’ resistant bacteria, for their own purposes. Surveillance thus consists in an arrangement of different spatialities, materialities and subjectivities (Enticott and Ward 2020) which allows the coexistence of several forms of relationship between humans, animals and microbes. However, the triple ontology of AMR that is enacted through the surveillance system (i.e. the three ontological referents that are produced by three types of engagement with microbes – those of microbiologists, epidemiologists, and veterinary practitioners) also engenders areas of uncertainty that limit the governability of resistant bacteria.
This chapter proposes an analysis of the history and functioning of Résapath,1 one of the apparatuses for monitoring resistant bacteria of animal origin. Designed to trace and control the flows of microbes and antibiotics within diverse animal populations, Résapath is France’s main AMR surveillance system. Its operation is based on the combination of several different tools and areas of knowledge, and in particular veterinary medicine, epidemiology and microbiology, which make it possible to collect bacteria and measure their levels of resistance to antibiotics. Résapath is organised like any other surveillance system in animal health. Veterinarians collect data during their routine animal care activities. For example, while on a farm visit or when a farmer calls them because an animal has fallen sick, veterinarians take biological samples to perform the tests required in the case of a bacterial infection (i.e. bacteria isolation and/or antibiotic susceptibility testing, also called ‘antibiograms’ – see the section ‘How does a surveillance system work’? in this chapter). These tests are processed by local laboratories which may either belong to the veterinary practice or be a private or public laboratory located relatively close to the farm. The results of these tests are sent both to the veterinarians, so that they can return to the farm and cure the sick animals, and to national veterinary epidemiology and microbiology units (usually belonging to expert bodies like food safety agencies). These units aggregate all of the data collected on the national territory and perform an in-depth analysis in order to monitor the global state of AMR (i.e. which resistances are increasing, which ones are decreasing, etc.). This analysis is then published in the expert agency’s annual reports and will inform the decisions of the national veterinary services (which are usually a part of a country’s Ministry of Agriculture) regarding which management strategies should be implemented to control the spread of AMR (e.g. a ban on or restriction of certain antimicrobials). The chart below (see Figure 8.1), which was adapted from the Manual of Livestock Disease Surveillance and Information Systems (FAO 1999), represents the functioning of surveillance systems in animal health.
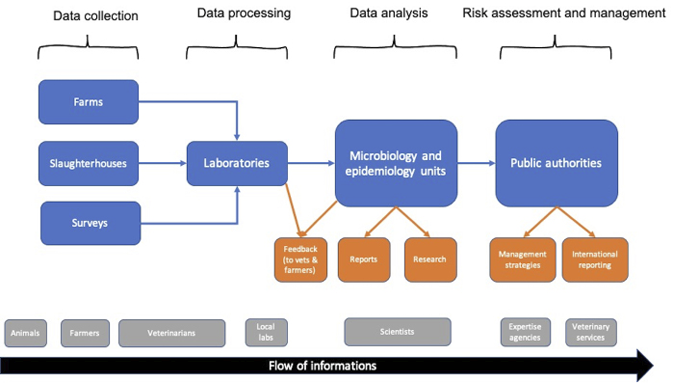
Fig. 8.1 Surveillance systems in animal health
Résapath is a network of member laboratories, distributed across France, which participate in the surveillance system and follow the corresponding protocols. It is coordinated by two national veterinary laboratories that are part of the French Food Safety Agency (ANSES), which oversees the epidemiology and microbiology units analysing the tests performed by labs on the samples submitted by local vets. One is based in Lyon and the other in Ploufragan (Brittany). Over the past 15 years, Résapath has grown considerably, with roughly 20 new labs joining the network and the number of antibiograms collected multiplying fivefold. In 2018, Résapath was composed of 71 local laboratories, mainly concentrated in the western area of France, and the network collected more than 55,000 results from antibiograms.
This chapter is based on two types of empirical material. First, I performed eight interviews with experts from the French Research Institute for Agriculture (INRA) and from the ANSES units in charge of Résapath, along with a dozen interviews with veterinarians who participate in the collection of the surveillance data. These interviews made it possible to understand the variety of practices relating to the surveillance activity. Second, we used a corpus of grey (activity reports, technical recommendations, annual reports) and scientific (veterinary epidemiology and microbiology articles, and transfer publications) literature produced by these actors, which allowed us to trace the evolution in the knowledge and techniques employed within Résapath since its creation, and even a few years prior to that. The chapter is divided into three parts and explores the role of microbiologists, epidemiologists and veterinarians in the surveillance of AMR, showing how their different engagements with bacteria tend to co-produce different ontologies of resistance.
Resistant bacteria: A job for microbiologists (the 1970s and 1980s)
Making AMR an object of research
The first people to study resistant bacteria of animal origin were veterinary microbiologists. However, although during the 1970s antibiotics were of interest to animal health researchers, the question of AMR was not yet a subject in its own right and continued to be split into different themes. Pathologists were interested in therapy, biologists were studying the effects of antibiotics as food additives, while pharmacologists were working on maximum residue limits. These areas of research finally discovered a point in common, AMR, thanks to a handful of microbiologists who decided to examine this phenomenon.
The researchers (Elisabeth Chaslus-Dancla, Jean-Pierre Lafont, and Jean-François Guillot) who were to bring this subject into the academic arena of veterinary microbiology shared the particularity of having received their training in places central to 1970s French microbiology, namely the team run by Professor Chabbert of the Pasteur Institute, and that of Professors Raibaud and Ducluzeau at INRA. The former was precisely where research into resistant bacteria was mainly to develop in France, although in relation to human medicine. The latter team was better known for specialising in the concept of ‘microbial ecology’ and for the techniques it was to develop throughout the 1970s, which would become an extremely important tool for AMR specialists.
The veterinary microbiologists set themselves up in a laboratory at INRA’s experimental station in Nouzilly, in the Loire valley. This station had a decisive feature that was to make it one of the essential nodes of the future surveillance system: Nouzilly owned an experimental chicken farm. This allowed the researchers to work on animal models that were not exclusively used to study human health issues (such as rats and mice) but which also (or even mainly) oriented studies towards animal health problems.
The team at Jouy was like Mecca, or the Vatican if you prefer, they were original thinkers, theorists on mice and rats. They worked on mammals, targeting humans, in Microbial Ecology. And at Nouzilly, we were closer to livestock. And chickens, they had the advantage of being both lab animals and livestock (INRA microbiologist).
However, having access to both laboratory animals and livestock was not the only key element. It was also because they were able to link this material to both the emerging theme of AMR (glimpsed during their time at the Pasteur Institute) and the microbial ecology techniques learned at INRA, that the Nouzilly veterinary microbiologists were gradually able to make resistant bacteria of animal origin a totally legitimate object of research, around and through which would be built an apparatus that would enrol a multitude of actors (human and non-human) into the ontological work of producing AMR. In the microbiology lab, humans and microbes interact in a specific way that makes this work possible. First of all, AMR was already a subject of academic research that was starting to flourish due to the development of molecular biology, which made it possible to study resistance genes. Second, the techniques of so-called ‘germ-free’ models, imported to France and developed by Raibaud and Ducluzeau, allowed them to work on fully sterilised laboratory animals. By inoculating these animals with bacteria that had predetermined characteristics (like resistance to antibiotics), humans were able to observe how microbes behave in contact with a change in their environment (e.g. after the introduction of an antibiotic molecule) without the experiment being flawed by any pre-existing and uncontrolled flora.
The creation of a bacteria ‘supply-chain’
At the beginning of the 1980s, the work carried out by the Nouzilly researchers was able to progress thanks to the creation of a socio-technical apparatus that enabled them to bring materials to their work bench. Microbiologists needed to regularly acquire bacterial strains with profiles (in this case, resistance) that were useful from an academic standpoint. Although there was little competition from their colleagues at the Pasteur Institute, who were not interested in bacteria of animal origin, the veterinary microbiologists needed their culture collection to be regularly renewed in order to ensure a certain frequency of publication. This was even more important given that at that time, their analytical techniques did not allow them to study the heart of the genome of resistant bacteria, which meant that they almost always needed a new strain in order to document a new resistance mechanism.
In 1982, Jean-Louis Martel, a veterinarian from the national veterinary laboratory in Lyon, created Résabo.2 This was a network of local veterinary laboratories collecting samples from field veterinarians. Initially, this network was anything but a surveillance system. In fact, it included less than a dozen laboratories, which sent poorly standardised results to the central laboratory in Lyon. Martel then requested that bacterial strains presenting an atypical resistance profile be sent to him so that he could study them in greater detail. Consequently, while the stated objective of Résabo was to produce data needed to measure the evolution of AMR in cows, its configuration made it no more than a network for circulating and supplying a limited number of bacterial strains for the purpose of microbiological research, and not of epidemiological surveillance.
As Martel’s laboratory did not really possess the skills and tools to carry out in-depth work on AMR, he began to collaborate with the Nouzilly team, sending them the most promising strains so that they could conduct a microbiological analysis of the resistance mechanisms. Starting from the 1980s, Martel was thus regularly associated with the publications made by INRA researchers. A division of scientific labour was then created, thus helping to structure the device, which at that time was organised through the link between Lyon and Nouzilly.
And for example, something that I did with Gérard [a PhD student in the mid-1980s] and that everyone now accepts, was to do with neonatal diarrhoea. […] I said it had to be the cow that carried it, but we had to prove it and we managed to do that via the initial resistances to quinolones, I remember a herd of cows that was monitored by a good practitioner and which had cases of diarrhoea. He was able to take samples from the mothers and the calves and sent the samples to Martel, who isolated the strains, so we had traceability that proved it (INRA microbiologist).
Two irreducible forms of knowledge? (the 1990s and 2000s)
The academicisation of microbiology
From the mid-1990s onwards, the operation of Résabo changed due to several dynamics that had an important impact on human-microbe relationships and therefore on the ontological co-production of AMR. First of all, the Nouzilly team experienced a process of ‘academicisation’ of their research, which meant their work was increasingly framed by the standards of high-level international science rather than by those of more applied research, which until then had been more typical of the work conducted at INRA (Tétart and Torny 2009).
The microbiologists’ engagement with animals and bacteria changed when a new researcher joined the team. He had already been working at Nouzilly’s experimental station but with a different team that was studying brucellosis, a zoonotic disease mainly found in cows and sheep. He was a microbiologist but not a veterinarian, a profile that prefigured the forthcoming recomposition of the team’s members. This researcher, who was to be put in charge of the laboratory a few years later, brought with him a new way of working that was far more oriented towards basic research (in particular, he used experimental mice models and introduced techniques taken from genetics). The team was gradually reorganised around works that were far more oriented towards high-level publications. Fewer bacteria were studied, but the research went much further in the genetic characterisation of resistance mechanisms.
This modification to the microbiologists’ engagement with resistant bacteria was also due to a change in their ‘supply-chain’. The new researcher had access to devices other than Résabo, enabling him to obtain more specific bacterial strains. In particular, he re-established the historical links with the Pasteur Institute and instigated collaborations with international teams. It was through these channels that he obtained a particular strain of salmonella (Salmonella DT104, known as ‘Kentucky’) which allowed the Nouzilly team to highlight a resistance mechanism that had enormous success in terms of scientific publication. From then on, the work of the Nouzilly microbiologists was organised almost exclusively around this singular mechanism (SGI1: Salmonella Genomic Island 1), thus strengthening the central position of these researchers within the surveillance system. The strictly academic engagement of the microbiologists within the apparatus helped produce a specific ontological referent for resistant bacteria, namely a bacterium-strain which was perceived individually and from a molecular and genetic perspective.
I identified a gene resistant to Florfenicol among the salmonellas. But when we looked a little bit around the genetic environment of this gene, we realised that it was an entire genomic island that carried other resistance genes. We ended up calling it SGI1 for Salmonella Genomic Island One. […] So, when we realised how important this was, it completely restructured the overall thematic of the research team, because it became the main study model, because we realised that it was a very good model for studying the propagation of antibiotic resistance genes (INRA microbiologist).
A short time after these developments at INRA, a second ‘historic’ member of Résabo also went through a form of academicisation in the early 2000s. When Martel retired in 2003, he was replaced by another veterinary microbiologist, albeit with a relatively different profile. The new director of the Lyon Bacteriology unit, who had previously cut his teeth with BSE,3 wished to raise the academic level of the team and change its role within the surveillance system. The stated objective was to no longer be a mere channel for supplying bacterial strains to INRA researchers.
Jean-Louis Martel recovered the veterinarian bacterial strains and sent them to INRA. It wasn’t a real partnership. It was a sort of a deal, a tandem that worked well like that. So Jean-Louis Martel was mentioned on INRA publications, never last, always somewhere in the middle. But he hadn’t really developed the same competency in his own laboratory. That’s what was missing (Microbiologist from ANSES-Lyon).
Such a change in the engagement of the Lyon microbiologists obviously took time (necessitating the appropriation of molecular biology knowledge and techniques, acquisition of material, targeted recruitments, etc.), but this academicisation was gradually achieved. While the Lyon-Nouzilly duo still constituted the heart of the apparatus, their relationship, the ensuing division of work, and above all the forms of engagement of its actors (human and non-human) underwent profound changes.
The emergence of the epidemiologists
The repositioning of the Lyon laboratory within the surveillance system was not solely due to this academicisation of the microbiology team. Résabo was significantly restructured between the late 1990s and the early 2000s under the impetus of the first French AMR policies.
First, Résabo merged with Onerba in 1997,4 thereby becoming part of a vast AMR surveillance system that had until then been exclusively comprised of networks dedicated to bacteria of human origin. This had a major impact on how it operated. The main challenge was the harmonisation of the ways in which data were produced, which was needed to ensure greater reliability. A standardisation process of data collection and processing was therefore carried out when Résabo joined Onerba, including the following: a charter of engagement and ‘good practices’ certification required for all local laboratories; a single data coding system; and regular training sessions. Moreover, the integration of the Lyon laboratory, along with all other national veterinary laboratories, into the new French food safety agency in the early 1990s also strengthened the work done to standardise the various epidemiological animal health surveillance networks, including Résabo (Dufour 1993). In 2001, Résabo became Résapath and was no longer limited to bacteria of bovine origin but could also collect and produce data from bacteria of porcine and avian origins. To this end, new local laboratories were gradually integrated and a second national veterinary laboratory (Ploufragan), specialising in the pig and poultry sectors, participated in the running of the network.
This restructuring had serious consequences for the forms of engagement within the apparatus. In particular, a new category of human actors emerged at this time: epidemiologists. These researchers quickly made AMR one of their preferred subjects and became highly engaged in the operation of Résapath. Since the emergence of the epidemiologists and the profound restructuring of the apparatus, the surveillance activity was at last able to develop. The epidemiologists’ engagement took forms that differed from those of the microbiologists and, above all, led to the production of another ontology of resistant bacteria. Alongside the bacterium-strain (enacted through an academic form of engagement), henceforth the apparatus also produced a bacterium-population (enacted through a surveillance form of engagement).
Although epidemiologists also engage in scientific research, their main aim is to produce a form of expertise that makes it possible to govern AMR. Therefore, unlike the microbiologists, they are not interested in bacteria as individuals, and even less in bacterial genetic and molecular mechanisms. They do not even have a lab where they can physically interact with bacteria or animals. They actually look at bacteria as a population and only need aggregated statistical data allowing them to trace changes within this population. Consequently, microbiologists and epidemiologists do not consider AMR in the same way. The former define a ‘breakpoint’, which is the threshold from which an antibiotic ceases to be effective on a given bacteria; the latter define a ‘cut-off’ which is the average point from which a population of bacteria ceases to be ‘wild’, i.e. it acquires some resistant genes, whether or not it is still susceptible to an antibiotic.
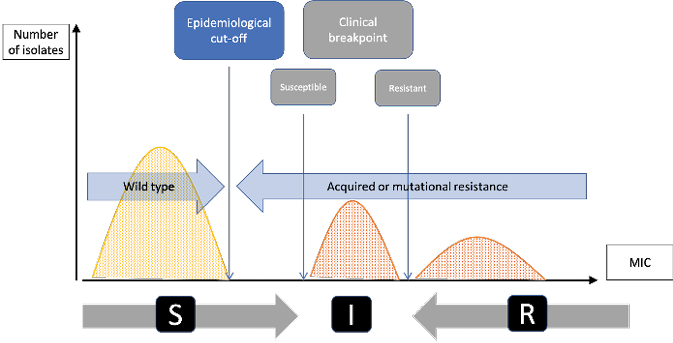
Fig. 8.2 Clinical breakpoints and epidemiological cut-off (reproduced from Tascini et al. 2016). This diagram shows the difference between the epidemiological cut-off (Ecoff) and the susceptibility (S) and resistance (R) breakpoints. Grey figures represent the microbiological conception of AMR: clinical breakpoints indicate likelihood of therapeutic success (S - susceptible) or failure (R – resistant) of an antimicrobial treatment against a given bacterial infection (I being the intermediary zone). Blue figures represent the epidemiological conception of AMR: Ecoff values separate microbes without (wild type) and with resistance. Figures are expressed in MIC which indicate the lowest concentration of a chemical (here an antibiotic) which prevents visible growth of bacteria. MIC is often expressed in milligrams per liter (mg/L).
How does a surveillance system work?
Back to the roots: How vets, animals, and microbes co-produce data
The work of microbiologists and epidemiologists, and the ontological referents of AMR which they produce, could never exist without some sort of an original engagement, i.e. an upstream sorting process that constitutes the basic material conditions of the surveillance system’s operation. Only the bacteria that Résapath has previously managed to ‘capture’ are likely to reach the microbiologists and epidemiologists. This first enrolment of non-human actors into the device is dependent upon another form of engagement and relationship with animals and microbes, namely that of veterinary practitioners who take the bacterial samples in order to produce the ‘antibiograms’ that the other two categories of actors need to obtain the data they require.
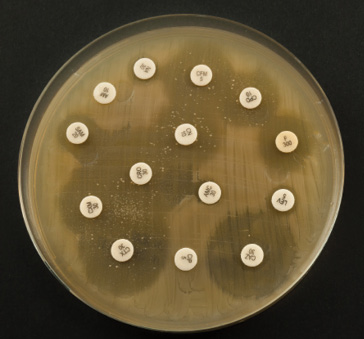
Fig. 8.3 An antibiogram (iStock, ref: 47080398). The antibiogram is the technical tool that makes it possible to characterise a bacterium’s antibiotic resistance profile. It consists of a petri dish that has been colonised by a bacterium. Using the sample that a veterinarian has taken from an animal, the lab staff isolate the bacterium (from the other cells and/or molecules present) and let it colonise the petri dish (generally no longer than 24 hours in an optimal environment). The actual antibiogram is then performed. A series of discs (the white dots shown in the picture) loaded with antibiotics are placed in the dish and one can observe how the bacterial colony reacts to each antibiotic: a large ‘inhibition diameter’ means that the bacteria is fully susceptible to a given antibiotic; an absence of diameter means that the bacteria is resistant since the antibiotic has no effect on its development.
Résapath is a so-called ‘passive’ surveillance network, meaning its existence is not subject to any regulatory obligation, and consequently its data are essentially produced through the routine activity of its field actors (Dufour and Hendrikx 2011). The way that veterinarians work with animals and microbes, and in particular the way in which they use (or not) antibiograms, is thus essential to Résapath’s operation. The results of the antibiograms are the apparatus’ base data which can then be used by the microbiologists, epidemiologists and even veterinarians according to their own specific objectives.
When a veterinarian takes a sample from a sick animal to determine the nature of the animal’s illness, he/she sends the sample to a local laboratory. The laboratory performs an antibiogram analysis to characterise the type of bacterium causing the infection and, at the same time, to determine its level of resistance to a certain number of antibiotic molecules. The laboratory returns the results to the veterinarian, who then decides how to treat the animal, and a copy of the results is sent to one of the two national laboratories running Résapath (assuming, of course, that the local laboratory in question is a member of the surveillance network). These national laboratories then do two things: the epidemiologists combine all of the collected data to calculate the global evolution in resistance levels for each bacterium(-population) in relation to each antibiotic, and the microbiologists may request that the bacterium(-strain) be sent to them if they believe its resistance profile is of interest. However, the possible existence of these two ontological referents is in fact dependent on the engagement of the veterinary practitioners without whom the system itself would not exist.
Veterinary medicine is above all a clinical activity, designed to care for animals. Veterinarians are therefore first and foremost interested in sick animals and, consequently, their pathogenic (rather than commensal) bacteria. The veterinarian’s ontological referent is thus a bacterium-disease (derived from a clinical form of engagement), which is significantly different from the microbiologist’s bacterium-strain (and academic form of engagement) or the epidemiologist’s bacterium-population (and surveillance form of engagement), which do not necessarily need to be pathogenic for their AMR profile to be of interest. There is therefore a selection process within Résapath’s operation which takes place through this primary engagement with microbes and animals; the only bacteria that can be captured are those that present a clinical interest for veterinarians. Indeed, veterinary practitioners only perform antibiograms when they are unable to treat sick animals, i.e. when they are in a situation of therapeutic failure. There is no sense in their using this relatively onerous and expensive protocol for infections which they can easily treat. Yet in fact, therapeutic failure usually means the failure of one or more antibiotic treatments. This means that the sick animals from which veterinarians take samples have already been treated with antibiotics and, therefore, a selection effect for the most resistant bacteria (those that resist the treatment) has already taken place before any enrolment of microbes into the apparatus.
Moreover, this selection effect relating to the clinical engagement of veterinarians with animals and microbes comes on top of the fact that veterinarians do not ask for an antibiogram every time they take a sample. Sometimes, veterinarians only want to obtain an ‘isolation’ from the laboratory, i.e. a characterisation of the bacterium implicated in the animal’s disease but not its resistance profile. This happens for relatively common infections for which they know that resistant bacteria are rarely found. In such situations, no data is produced for Résapath.
For a mastitis, [the veterinarians] can easily say: ‘I want to give the right treatment but there’s very little resistance with mastitis, I just want to know what germ it is, so I can use the right antibiotic. I don’t need to do an antibiogram because mastitis germs are quite susceptible, but I want to know if it’s a Coli or a Strepto or a Staph’. So they just ask for an isolation. And if there’s no antibiogram, we don’t collect anything (ANSES-Lyon epidemiologist).
The triple ontology of AMR: Complementarity and uncertainty
All in all, we can see the extent to which the clinical engagement of veterinarians is a determining factor of not only the knowledge but also the governability of resistant bacteria. Résapath is the socio-technical apparatus that gives life to the boundary-object of AMR by enrolling a wide range of actors which can enact it. However, this co-production of AMR is the result of heterogeneous forms of bonds and engagement between humans, animals and microbes that are both complementary and contradictory. There is an incommensurability between the ontological referents produced by the Résapath apparatus that can engender uncertainty in the governance of AMR.
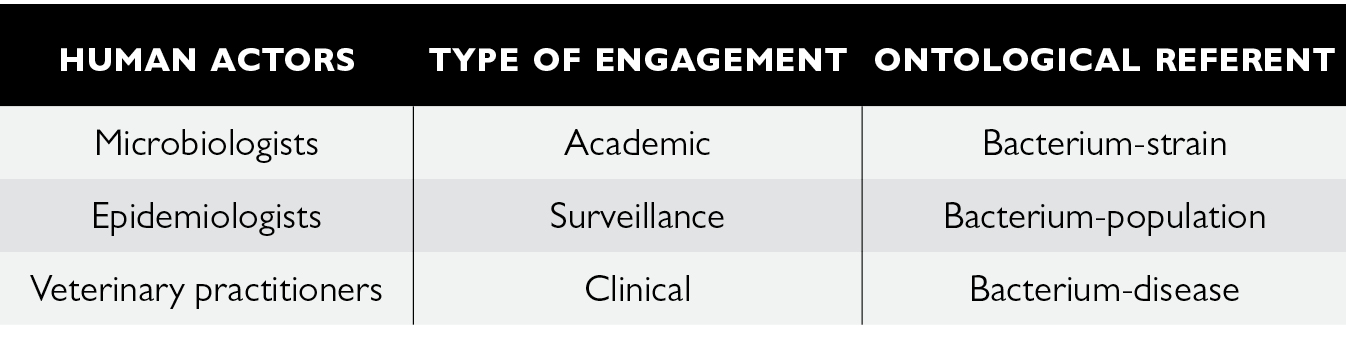
Fig. 8.4 The triple ontology of AMR enacted through Résapath
By assembling a range of human and non-human actors whose engagement is not based on the same objective (be it clinical, microbiological or epidemiological), the apparatus produces competing definitions of AMR which make it difficult to entirely fulfil any purpose. Although the development of Résapath has been a great provider of bacterial strains of interest for microbiologists for two decades, the stabilisation of the system (in terms of territorial extension, sampling practices, analytical tools, etc.) makes it less likely now to regularly capture original bacteria and renew the strain bank of microbiologists. Veterinary practitioners cannot fully rely on antibiograms as they are unable to predict with complete certainty the therapeutic effectiveness of antibiotics (Fortané 2015). Epidemiologists combine and model data whose statistical representativeness may be questioned (Botrel et al. 2006), as they remain dependent upon the decisions of veterinarians regarding whether or not to perform antibiograms (Bourély et al. 2018).
So, we’re talking about a sort of mixture, in fact it’s written on the veterinary CA-SFM5, at the top it’s written ‘the data are of an epidemiological nature’, because we’re really looking to see whether it’s resistant or not resistant on the diameter distribution curves. Of course, we can’t judge the clinical effectiveness for vets because we don’t even know if it works or not, in other words at the end of the day whether the cow is alive or dead. We have no idea. So, we’re in a clinical frame of reference but with epidemiological data. (…) So, it’s a little bit this ambiguity which means that the surveillance is nevertheless linked to a system which, initially, was to help the practitioner. So, it’s an in-between thing (CA-SFM microbiologist).
As shown by this interview, the triple ontology of resistant bacteria engenders something that is ‘in-between’, which I relate to a form of ungovernability in the sense that none of the three human-microbe relationships can be entirely fulfilled. Governing microbes does not only refer here to AMR policy but principally to the assemblage of practices and engagements with microbes through which AMR is enacted. Even if these multiple enactments are (and need to be) partially combined since they rely on each other, several ontologies of resistant bacteria keep co-existing and producing some sort of ‘in-between’ governability of AMR where every human actor has to deal with a share of uncertainty. Because of the way Résapath works, neither veterinarians (who must adjust and control the antibiotic treatment of sick animals), microbiologists (who must set adequate therapeutic thresholds for each resistant strain), nor epidemiologists (who must monitor the status of the bacterial population) can fully rely on the knowledge that is crucial for the accomplishment of their objectives.
In the end, what do we know and govern about AMR? Is it the evolution of a bacteria population or the presence of resistant genes in a given environment? Is resistance the individual ability of a bacteria to definitely inactivate an antibiotic’s effect, a genetic evolution that alters the susceptibility to an antibiotic until the pharmacological structure of this antibiotic can be adapted to this evolution (for example by increasing the MIC), or just a momentaneous feature of a bacterial population that may naturally disappear anyway if we reduce its exposure to antimicrobial compounds? It is probably a little bit of everything, and this is what makes AMR so complex and uncertain, and in some ways so controversial. Yet reducing the use of antibiotics in both human and veterinary medicine is undoubtedly an important goal for managing the risks posed by resistant bacteria. However, the relationships between humans, animals and microbes are certainly richer and more diverse than a surveillance system could ever capture, even though its operation and efficiency already rely on a combination of different epistemologies and ontologies.
Conclusion
From a small network of a dozen or so local veterinary laboratories in 1982 to a system that now comprises around 70 laboratories producing 55,000 antibiograms a year (compared to between 10 and 20 times fewer 40 years ago), Résapath’s socio-technical organisation has undergone significant changes. The surveillance of resistant bacteria of animal origin is now based on the interlinking of several different forms of engagement and relationship between humans, animals and microbes, and produces three ontological referents for resistance: the veterinarians who collect (and therefore select) the bacteria in accordance with their professional motivations (in particular that of caring for animals) are clinically engaged in the apparatus and co-produce a bacterium-disease; the microbiologists who are above all looking to get material onto their lab bench with new microbial strains are academically engaged in the apparatus and co-produce a bacterium-strain; and the epidemiologists who aim to model the evolution in bacterial resistances to antibiotics are engaged in the apparatus for surveillance purposes and co-produce a bacterium-population. All in all, surveillance arranges different spatialities, materialities and subjectivities in order to enact the (multiple) existence(s) of AMR.
Additional research would nevertheless be useful since in this chapter I have only examined one type of apparatus – that of an AMR passive surveillance system. Yet there are also active surveillance networks (such as salmonella monitoring in slaughterhouses), systems for post-market authorisation surveillance (monitoring the side effects of pharmaceuticals) and for the monitoring of antibiotic sales (data from the pharmaceutical industry), prescription (data from veterinarians), and use (data from livestock farmers). It is likely that these apparatuses produce other ontological referents because they mobilise different instruments, knowledge and metrics, particularly those from the field of pharmacology, which has other ways of measuring and defining AMR, such as pharmacokinetic and pharmacodynamic models. Nor should we forget that resistance, while a property of bacteria, is first and foremost the result of interaction between the latter and antibiotic molecules in specific social, biological and ecological contexts (human and animal bodies or the environment, including farms, healthcare facilities and sewers). Yet all of these organisms are unstable: pharmaceutical companies regularly update the composition of their antibiotics in order to improve their effectiveness and safety (in particular by relying on data from all of these monitoring and surveillance systems); the food industry is continuously transforming animals’ bodies (genetic selection); environments are constantly subject to pressure from phenomena such as climate change or the discharge of chemical substances into the water, air and soil. There is no doubt that several resistance ontologies exist, and if it seems so difficult to govern AMR, this is perhaps also due to the fact that AMR is so many different things at the same time.
Notes
1 From the French: Réseau d’EpidémioSurveillance de l’Antibiorésistance des bactéries PATHogènes animales (epidemic surveillance of antibiotic resistance from pathogenic animal bacteria).
2 Network for the epidemio-surveillance of the antibiotic resistance of pathogenic bacteria of bovine origin.
3 Bovine spongiform encephalitis, commonly known as ‘mad cow disease’.
4 National observatory for bacterial resistance to antibiotics.
5 Antibiogram committee of the French Society of Microbiology. This is the expert body that sets up and reviews antibiogram thresholds annually.
References
Botrel, M.-A., and others, ‘Le Résapath: analyse critique et propositions d’amélioration’, Epidémiologie et santé animale, 50 (2006), 157–68.
Bourély, C., and others, ‘Why Do Veterinarians Ask for Antimicrobial Susceptibility Testing?’, Preventive Veterinary Medicine, 159.1 (2018), 123–34.
Callon, M., ‘Éléments pour une sociologie de la traduction. La domestication des coquilles Saint-Jacques dans la Baie de Saint-Brieuc’, L’Année sociologique, 36 (1986), 169–208.
Doré, A., ‘L’exercice des biopolitiques. Conditions matérielles et ontologiques de la gestion gouvernementale d’une population animale’, Revue d’Anthropologie des Connaissances, 7 (2013), 837–55.
Dufour, B., ‘Naissance et développement de l’épidémiosurveillance animale en France’, Epidémiologie et santé animale, 23 (1993), 83–100.
Dufour, B., and P. Hendrikx, Surveillance épidémiologique en santé animale, 3rd edn (Paris: Editions Quae, 2011).
Enticott, G., ‘Navigating Veterinary Borderlands: “Heiferlumps”, Epidemiological Boundaries and the Control of Animal Disease in New Zealand’, Transactions of the Institute of British Geographers, 42.2 (2017), 153–65.
Enticott, G., and K. Ward, ‘Mapping Careful Epidemiology: Spatialities, Materialities, and Subjectivities in the Management of Animal Disease’, The Geographical Journal, 186.3 (2020), 276–87.
Food and Agriculture Organisation (FAO), Manual of Livestock Disease Surveillance and Information Systems (FAO, 1999).
Fearnley, L., ‘Epidemic Intelligence. Langmuir and the Birth of Disease Surveillance’, Behemoth, 3 (2011), 36–56.
Fortané, N., and K. Keck, ‘How Biosecurity Reframes Animal Surveillance’, Revue d’Anthropologie des Connaissances, 9.2 (2015), a–l.
Fortané, N., ‘La triple ontologie des bactéries résistantes’, Revue d’Anthropologie des Connaissances, 9.2 (2015), 265–90.
Kirchhelle, K., Pyrrhic Progress: The History of Antibiotics in Anglo-American Food Production (New Brunswick: Rutgers University Press, 2020).
Lakoff, A., and S. Collier, eds, Biosecurity Interventions. Global Health and Security in Question (New York: Columbia University Press, 2008).
Lakoff, A., ‘Two Regimes of Global Health’, Humanity: An International Journal of Human Rights, Humanitarianism, and Development, 1 (2010), 59–79.
Law, J., and A.-M. Mol, ‘Veterinary Realities: What Is Foot and Mouth Disease?’, Sociologia Ruralis, 51.1 (2010), 1–16.
Manceron, V., ‘Recording and Monitoring: Between Two Forms of Surveillance’, Limn, 3 (2013), 25–30.
O’Neill, J., ‘Tackling Drug-Resistant Infections Globally: Final Report and Recommendations’ (final report of the Review on AMR commission, London, UK, 2016).
Star, S., and J. Griesemer, ‘Institutional Ecology, “Translations”, and Boundary Objects: Amateurs and Professionals on Berkeley’s Museum of Vertebrate Zoology’, Social Studies of Science, 19.3 (1989), 387–420.
Tascini C, Sozio E, Viaggi B and Meini S, ‘Reading and understanding an antibiogram’, Italian Journal of Medicine, 10 (2016): 289–300.
Woolgar, S., and J. Lezaun, ‘The Wrong Bin Bag: A Turn to Ontology in Science and Technology Studies?’, Social Studies of Science, 43 (2013), 321–40.