4
Sensing Salmonella: Modes of sensing and the politics of sensing infrastructures
Francis Lee
Entering the ECDC
Entering the European CDC (ECDC) in Stockholm.1 It’s a sunny day in January. I’m heading for the first day of fieldwork. On the doors to the ECDC, and throughout the building, signs are posted that declare ‘Threat Level 0’. I’m given a badge, an ECDC laptop, and a desk in the Operations Centre, the set of rooms that can be claimed for intense operations, such as during the Ebola crisis. I soon learn that it is mostly used for routine work and meetings in the interim. That’s why I can borrow a table there. The Operations Centre consists of several rooms, the most important being the ‘situation room’, which is set up just as the classic image from the movies: A large table with perhaps 20 spaces, a wall of screens showing world news, a few tables with telephones for operators under the screen-wall, and of course a blinking red digital clock that shows the time in Stockholm, Atlanta, Brasilia, and Beijing.
During my fieldwork, every weekday at 11:30, I join about twenty experts from across the ECDC for the daily roundtable meeting – to assess the current disease threats against European citizens. During my fieldwork both mundane and exotic threats were part of the bestiary of threats: seasonal Flu, Zika, Legionella, Salmonella, Yellow Fever, and Plague were all brought under the scrutiny of the roundtable. Recommendations for action were produced. Debates about the right course of action were common. As I came to understand disease surveillance better, I came to think of this room as one of the central locations where disease outbreaks were sensed and where the right course of action was decided.
How do we analyse the politics of sensing infrastructures?
The intent of this chapter is theoretical and empirical. First, theoretically, the chapter proposes an analytical concept, modes of sensing, that is intended to examine how sensing infrastructures become implicated in the politics of sensing. The point is that attending to conflicts between different modes of sensing allows the analyst to become sensitive to differences, oppositions, and hierarchies between sensing infrastructures. The argument is that if sensing infrastructures are linked with different politics, then we also need analytical tools that allow for the description and analysis of oppositions, hierarchies, and indeterminacies that arise between different sensing infrastructures – and the politics that these differences in sensing give rise to and are implicated in.
Second, empirically, the chapter analyses how different sensing infrastructures create different understandings of what an epidemic is, where it originates and develops, and what its essential properties are. In essence: how different modes of sensing constitute disease outbreaks in different manners. Thus, the chapter sketches how an emerging sensing infrastructure for disease surveillance – in this case a sensing infrastructure based on genetics – becomes both championed and contested as evidence of a disease outbreak. It analyses how the introduction of a new infrastructure for sensing disease leads to the performance of a new disease object in Europe – a ‘long ongoing trans-European outbreaks of Salmonella’ as my informants would have it – and how actors at the ECDC and elsewhere struggle to reconcile this genetically detected outbreak with other modes of sensing disease: Does the disease outbreak originate from Country X or not?2 Which sensing infrastructure becomes dominant? And with what consequences?
Concretely, the chapter analyses how actors at the ECDC handle the uncertainties involved in the introduction of this genetic sensing infrastructure, and the actors’ work to coordinate and handle conflicts between the emerging genetic sensing infrastructure and more entrenched ways of sensing disease outbreaks. To achieve this, the chapter traces how the emerging genetic sensing infrastructure and entrenched ways of sensing disease are tied to different modes of sensing, that sometimes diverge and thus must be coordinated in practice. That is, the introduction of the genetic sensing infrastructure and the detection of a new class of outbreaks based on this infrastructure leads to practical, epistemic, and political tensions that need to be negotiated organizationally and politically. The chapter thus proposes a practice-oriented analysis of the politics of sensing.
An important facet of disease surveillance, which also makes it a particularly fertile ground for analysing the politics of sensing infrastructures, is that disease surveillance is fraught with politics of the most mundane kind – something which is also quite apparent in this time of the Covid-19 pandemic. Apart from the obvious health consequences of a large disease outbreak, the social and economic repercussions can be momentous. For instance, an outbreak can hinder tourism, it can stop the import or export of foodstuff or even topple politicians. Due to the potentially large consequences of disease outbreaks, conflicts about the origin of a disease, or the handling of an outbreak, can erupt between different national governments, as well as different types organizations and companies. For instance, the fear of a Zika pandemic became a global controversy leading up to the Rio Olympics. The purported discovery of Zika-cases in Tanzania led to the sacking of the director of the national institute for medical research. And a Russian ban on the import of German cucumbers due to an E. Coli outbreak lead to an international row.3 Thus, disease surveillance implicates national governments, private companies, as well as international organizations in a constant quest for surveilling and preventing new disease outbreaks – as well as conflicts around their detection and the possible political repercussions.
Disease surveillance, biosecurity and a tidal wave of sensing technologies
Current research dealing with disease surveillance in the social sciences has mainly been focused around the concept of ‘biosecurity.’ This body of work, pivoting around broad Foucauldian and anthropological perspectives, often analyses how biosecurity is handled by a diverse array of experts, on the scientific, political, and social levels. The analytical thrust of this body of work is aimed at understanding the institutional structures of expertise, and the construction of a multitude of objects of knowledge. The body of work paints a picture of a new world of constant preparedness, or in some cases unpreparedness, against the next global pandemic (Lakoff and Collier 2008; Lakoff 2017; Caduff 2015; Keck 2010).
While this research on biosecurity highlights the need for wide-ranging cultural, institutional, scientific, and political analyses of disease surveillance, what is frequently missing from this line of inquiry is an interest in the intertwining of disease surveillance with technological devices and infrastructures. Different disease experts, laboratories, and organizations are continuously attempting to harness new technological infrastructures aiming to find new ways of sensing disease: genetics, search word analyses, data-mining, machine learning, disease modelling, and risk computations are among the technologies that are mobilized to track and surveil disease globally (cf. Lee 2021).
Thus, an important point of departure for this chapter is that today’s disease surveillance is dependent on a varied array of information infrastructures. The importance of information infrastructures for the construction, classification, and acting in the world has been a classic topic in Science and Technology Studies (STS). Bowker and Star (1999), for instance, have crucially shown how infrastructures can impose a ‘social and moral order’, and have argued for an analytical strategy of infrastructural inversion, which ‘means learning to look closely at technologies and arrangements that, by design and by habit, tend to fade into the woodwork [and] recognizing the depths of interdependence of technical networks and standards, on the one hand, and the real work of politics and knowledge production on the other’ (Bowker and Star 1999: 34). Thus, Bowker and Star called for and instigated a wide-ranging ethnographic, historicizing, and practice-oriented, engagement with the negotiated and complex politics of infrastructures (see Star and Ruhleder 1996; Bowker 2000).
In today’s society where algorithms, machine learning techniques, and big data are constantly reshaping society in a multitude of ways information infrastructures are becoming increasingly important for our understanding of the world. Observers have for instance highlighted that new ‘Big Data’ infrastructures – and ways of knowing the world – will lead to new paradigms in how we create knowledge and facts (Kitchin 2014; Boellstorff 2015). Others have highlighted how computer algorithms or machine learning become part of valuing, classifying, and performing the world (Lee 2021; Lee et al. 2019; Lee and Björklund Larsen 2019; Seaver 2017; Kockelman 2013; Ziewitz 2017; Mackenzie 2017). Furthermore, what has also become apparent in this technological moment is that it is not only the amassing of great amounts of data, or the analysis of this data through different computational means, have exploded, but there is also a torrential downpour of new sensing devices and technologies. These sensing infrastructures draw on many different types and sources of data. For example, social media tracking, computer models to predict risks, satellite data, as well as a plethora of algorithms and computer software to make sense of this tidal wave of data (cf. Lee 2021).4
In disease surveillance, the emergence of new sensing infrastructures has the potential to make new disease outbreaks detectable. In other words, new classes of outbreaks and disease risks can be detected and made into objects in society through the development and introduction of new sensing infrastructures. For example, through satellite imaging and environmental computation, disease surveillance organizations can make environmental predictions of where different disease vectors could thrive on a global scale. A type of global analysis which was previously impossible (cf. Lee 2021).
Analysing sensing infrastructures and the politics of sensing
In attending to this technological moment of exploding sensors and sensing infrastructures, Gabrys (2016) has pointed out that new entities or environments are constructed, in her parlance concresce, through different sensing infrastructures. An important point being that different ways of sensing the world constructs it in different ways, with large consequences for what type of ‘politics … take hold along with these technologies’ (18). This means that ‘new modes of … data gathering’ lead to ‘new configurations of … engagement, …relationality, sensing, and action’ (23). The impetus of Gabrys’ work thus opens up a space for analysing and reflecting on sensors as linked to different politics of sensing.
Gabrys’ point that sensing infrastructures are linked to new configurations of engagement and politics is also true for disease surveillance. New sensing infrastructures lead to new disease outbreaks being sensed, and these new instances of disease outbreaks lead to new ways of engaging with, relating to, and acting on disease. The thrust of Gabrys’ work thus points to a need for engaging with how disease surveillance deals with these emerging infrastructures and technologies. However, Gabrys’ work also begs the question of how to deal analytically with these different infrastructures and politics of sensing? How do we move from the insight that different sensing infrastructures are linked to different types of politics, to analysing the politics of sensing infrastructures?
To make things more concrete: for instance, utilizing web searches in order to track the flu is not fully trusted as evidence of flu outbreaks in the Swedish healthcare system; rather people concerned with tracking the flu wish to rely on other types of sensing disease, such as lab reports or sentinel-reporting. In this situation, different modes of sensing flu intensity, through different sensing infrastructures, need to be coordinated. If the two sensing infrastructures diverge in making a ‘flu epidemic’, which one then becomes dominant?
Modes of sensing: Analysing a multiplicity of sensing infrastructures
To analyse these politics of sensing infrastructures this chapter introduces the concept of mode of sensing.5 This concept highlights not only the emergence of different infrastructures and politics of sensing, but also the constant multiplicity of sensing infrastructures in practice, and the political struggles that can emerge from concurrent uses of different sensing infrastructures. The point is to highlight how multiple sensing infrastructures and modes of engaging with the world need to be handled in concurrent situations, such as in the case of the flu epidemic alluded to above. But how do we approach this multiplicity of sensing infrastructures analytically?
Coordinating multiple sensing infrastructures
In Mol’s (2002) well-known analysis of the multiplicity of disease she traces how one disease, atherosclerosis, is made – enacted – differently in different parts of a hospital, sometimes in incommensurate manners. In this analysis, Mol attends to how different versions of atherosclerosis, different versions of this particular object, are handled in hospital practice. She attends to how different versions of the disease are coordinated, distributed, and included in each other. That is, she pays attention to how the different enactments of an object in different parts of the world are in need of constant coordination to become a coherent object. Thus, what Mol’s approach highlights is how objects are constantly made in in different manners, and that there is a constant need for coordinating the different versions of objects in practice.
Mol’s argument about the multiple enactments of objects resonates with Gabrys’ focus on how the world concresces differently around different sensing infrastructures. Just as Mol’s atherosclerosis is made differently in different places in the hospital, Gabrys’ environment is made differently with different sensing infrastructures. However, Mol’s focus on the coordination of multiple versions of objects also points to ways in which Gabrys’ analysis of sensing infrastructures can be developed to become sensitive to simultaneous and multiple enactments. If we are currently living in a veritable flood of sensors and sensing infrastructures, Mol’s perspective can be used to call attention to how objects and worlds, are overdetermined by multiple sensing infrastructures. Thus, Mol’s perspective highlights how Gabrys’ work on sensing infrastructures can be extended and highlights the need for analysing the coordination of multiplicities of sensing infrastructures.
Modes of sensing: Maintaining an analytical focus on sensing infrastructures
The introduction of the concept of modes of sensing is consequently intended to show how multiple and different sensing infrastructures are coordinated. Just like in Mol’s work, different enactments of objects, based on different modes of sensing, can both co-exist or clash. However, unlike Mol’s work which focuses on the multiple practices of enacting objects, the concept of modes of sensing intends to highlight the work of handling and coordinating different infrastructures. Thus, by introducing modes of sensing my intention is to direct our attention toward the politics of sensing, and the infrastructures that makes sensing possible.6
Consequently, the introduction of modes of sensing is an infrastructural inversion of Mol’s work (see Bowker and Star 1999: 34). That is, Mol’s focus on enactment, or the making of objects in multiple practices, highlights the simultaneous unity and multiplicity of objects in practice, and the need for coordinating different versions of objects. However, Mol’s work does not systematically engage with infrastructures and the question of whether particular modes of sensing come to dominate over others. In other words: In Mol’s work there is no systematic attempt to analyse how different knowledge infrastructures fit with larger struggles about what becomes the dominant enactment of the object.7
Modes of sensing Salmonella
Thus, in this chapter, it is the constant negotiation and coordination between different infrastructures that is highlighted. The focus is on the politics of different modes of sensing. Which mode of sensing Salmonella becomes dominant? And in which situations? This allows an analysis of the politics of sensing in practice – and, in this case, the politics of sensing disease outbreaks.
In sum, to explore the coordination and multiplicity of sensing infrastructures – and the enactment of Salmonella in Europe – this chapter develops the observation that there are different modes of sensing the world. The argument is that an analytical attention to modes of sensing allows us to describe how different sensing infrastructures clash or cohere. I suggest that an analysis of sensing infrastructures benefits from paying attention to multiple modes of sensing, as well as hierarchies between different sensing infrastructures. This is the point of departure for this chapter, and the basis for introducing the concept of mode of sensing.
In the present case – of a contested European Salmonella outbreak – two different sensing infrastructures can be tied to two different modes of detecting disease outbreaks, with potentially large political, economic and organizational consequences. An important point being that the same disease objects – the same epidemics – can be enacted in different manners with different sensing infrastructures, and that these divergent ways of sensing disease need to be handled in practice.8
Methodology
The chapter builds on fieldwork done for a larger research project that examines how new infrastructures are reshaping disease surveillance. The project started in 2015 with a preparatory inquiry into the rise of ‘infodemiology’, that is, how new information infrastructures and new types of data are harnessed for the purposes of disease surveillance. These new infrastructures can for example entail genetic data, web searches, tweets, sales data, or travel information.
The material for this particular chapter draws on my fieldwork in the spring of 2017 at the European Center for Disease Control and Prevention (ECDC).9 The fieldwork entailed three weeks of intense participant observation in the so-called epidemic intelligence team, follow up visits to observe the genetics team, as well as interviews and document analysis. Access to the field was granted after initial contacts with the team leader for the so-called epidemic intelligence team. The epidemic intelligence team is tasked with trawling social media, news media, and a constant flow of emails, and it reports to produce a snapshot of the current disease state of the world, while the genetics team uses genetic profiling of different organisms to surveil and trace disease.
During my fieldwork I did surveillance work, attended meetings, participated in staff training, and interviewed my informants formally and informally. During my stay I was free to attend meetings and training sessions with the epidemic intelligence group, as well as with other groups. The backbone of the surveillance process – as well as my understanding of disease surveillance at the ECDC – was the daily roundtable meeting where different teams from the ECDC brought the current day’s disease threats for assessment. Thus, the current chapter draws on participant observation, informal conversations, interviews, working documents, flowcharts, official ECDC publications, as well as online material. During the fieldwork meetings, conversations, and interviews were conducted and documented in field-notes. After the fieldwork, the chapter was complemented with some informal interviews, and emails as well as complementary document studies.
Importantly, in many cases disease surveillance at the ECDC is a political balancing act. The ECDC is located in the complex setting of European bureaucracy, where different agencies and bodies of government have different responsibilities. This means that the ECDC must navigate a complex organizational role where national governments and different public health agencies of the EU member states must be taken into account. For example, the ECDC does not act, it only monitors disease. It then reports these disease threats in a steady stream to the European commission and member states, which then decide how to act. All of these organizational entanglements have consequences for the practices and results of disease surveillance. In the case of Salmonella, which is the disease with which we deal in this chapter, a crucial part of the organizational puzzle is the EFSA, the European Food Safety Authority, which is responsible for handling food safety issues in the EU.
As a consequence of doing fieldwork in a particular location, the chapter is written from the point of view of a partial and situated knowledge of disease surveillance practice. To create a more comprehensive understanding of disease surveillance would entail following disease security practices not only at the ECDC, but at the public health agencies of various European member nations, in various laboratories in different countries, as well as in different European organizations. The chapter must therefore remain a locally situated intervention, into a local enactment of a particular disease outbreak. This approach of course limits the amount of data that is available from national authorities on the Salmonella outbreak in ‘Country X’, as well as how other European organizations, such as the EFSA, understand and enact this particular Outbreak of Salmonella. However, through observations, interviews, and document studies, the chapter attempts to sketch how different sensing infrastructures and different modes of sensing are handled in different situations.
Shoe leather epidemiology
In disease surveillance the origins of disease is a matter of big concern. Ever since the iconic work of John Snow – who traced the origins of the London cholera epidemic in the nineteenth century – disease surveillance has been focused on tracing the origins of disease through an eclectic combination of detective work based on any available methods of tracing and tracking disease. Snow, for example, produced a map of disease cases that allowed him to deduce the location of the source of the London cholera epidemic. To accomplish this, he drew on medical theories, knowledge of the local neighbourhood, as well as what ECDC natives sometimes term ‘shoe leather epidemiology’ – lots of walking, talking, looking, and thinking.
The traditional way of sensing Salmonella has been dominated by exactly this type of shoe leather epidemiology – tracing foodstuffs through their journey from farms, through production facilities, stores, and restaurants, all the way to the mouth of the European Citizen. Trying to find the restaurant where the disease was spread, the wedding reception where the bad eggs were used – or in the politically most momentous cases – the factories and industrial production facilities where disease is circulating. That is, the traditional shoe-leather tracing of Salmonella depended on tracing foodstuffs from their consumption to a potential contaminant. This process entails tracing foodstuffs through a network of producers, distributors, and retailers. If shoe leather evidence can be secured, the localized outbreak might be traced back to a certain farm or factory.
Of course, this shoe leather work also draws on large infrastructures of food traceability. Packages are marked, shipments are enumerated, lots are numbered. Tracing food from farmstead to mouth. These infrastructures make easier the detective work of tracing disease but should the package or the eggshells already be disposed of, the work of tracing origins becomes much, much harder. If not impossible.
This mode of sensing Salmonella depends on linking specific disease cases through food networks, to hopefully find the source of disease. Two cases can only be linked to each other if both cases can be traced back to the source. There is no way of saying that these share an origin story without linking them by tracing eggshells, food containers, or shipment lots to a certain farm, factory, store, or restaurant. However, this mode of sensing of Salmonella is changing through the introduction of new genetic technologies.
Genetics: An emerging sensing infrastructure
Disease surveillance practitioners are constantly experimenting with different sensing infrastructures. Oftentimes, resorting to any means possible to track down and eliminate the sources of a disease. During my fieldwork I came across instances of using TripAdvisor to find the location of outbreaks, analysis of satellite imagery to track climactic suitability for different disease vectors, machine learning techniques to model the spread of disease vectors, and news trawling to find new outbreaks of unknown diseases. New technologies that become available are constantly experimented with and can range from genetics to twitter analysis.
One of the emerging and promising trends in disease surveillance at the time of my fieldwork was harnessing affordable so-called whole genome sequencing (WGS) for purposes of disease surveillance. The affordability of WGS was time and again described as a breakthrough for tracking and tracing disease during my fieldwork. I argue that it can be productively understood as an emerging sensing infrastructure in disease surveillance.10 For instance, when I attended the ESCAIDE 2016 (European Scientific Conference on Applied Infectious Disease Epidemiology), which gathers hundreds of disease surveillance specialists from all over Europe, genetic tracking of disease was one of the dominant themes.
As the head of disease surveillance at the ECDC expressed it in an informal conversation during my fieldwork: genetic surveillance heralded the future of disease surveillance.11 Another of my informants, the head of the genetics team at ECDC, reflected in a personal communication on how WGS is changing how disease surveillance is done:
Traditionally outbreak detection has been built up of two parts. One is the epidemiological link [“shoe leather epidemiology”] where certain food can be suspected because of evidence or consumption of specific [food] items between cases. [Previously] this has […] been complemented with some crude laboratory methods to conclude that the same bacterial strain in present in between cases and hopefully also [the] food item.
Now the weight of these two pieces of evidence is tilting, because you get so detailed and high resolutive microbiology data [from genetics]. Earlier the question was, how much laboratory data do you need to conclude a source based on epidemiological evidence. Now the question is reversed to how much epidemiological evidence is needed to conclude a link from something detected by genomics (personal communication to author).
What my informant was pointing out is how the advent of an emerging sensing infrastructure based on genetics leads to shifts in how disease outbreaks are understood and detected in practice. He also points out how different sensing infrastructures are trusted differently. From his situated point of view, as the head of the genomics team at the ECDC, there has been a reversal in how evidence from different sensing infrastructures is trusted. However, as this chapter shows, the introduction of a genetic sensing infrastructures is not as smooth as it might appear from the point of view of the genetics team. Below we follow the tracking of a particular outbreak of Salmonella, and the work of the genetics team to attempt to find a source for this particular outbreak. We follow how the emerging genetic sensing infrastructure is implicated in a political and organizational conflict between two different enactments of a salmonella outbreak.
Genetic epidemiology and phylogenesis
With the advent of affordable so-called whole genome sequencing, and the drive to use genetics to track disease, the shoe leather way of enacting Salmonella outbreaks is changing. Now, rather than tracing eggshells or packages through the food chain, disease is starting to be traced through genetic similarities of strains of bacteria. This work builds on the logic of genetic similarity and difference, where relations between strains of bacteria are inferred by genetic closeness. This logic of genetic similarity and relation is perhaps most clearly expressed through so-called phylogenetic trees, where the ancestries of species are drawn in tree structures based on changes in the genetic code. The theory of phylogenesis is based on an evolutionary logic where changes in the genome give rise to genetic differences, and in the end new species.
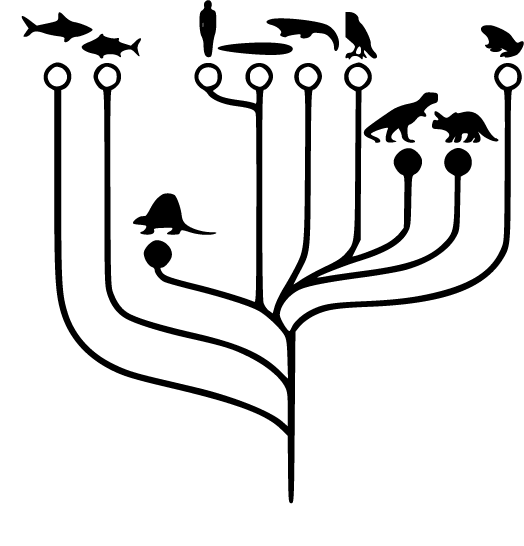
Fig. 4.1 Phylogenetic tree (source: redrawn from Fig. 5 in Carlson 1999)
The logic of introducing phylogeny into surveillance is based on theories both about how evolution happens, but also on how species come to be differentiated genetically. As one of my informants phrased it: ‘The fact is that evolution is constantly diversifying organisms and this can be visualized and applied practically.’12 When creating phylogenetic trees through genetic studies, genetic similarity is often equated with a close relation between particular species or organisms. There is thus a general figure of thought in studies of phylogenetics, where genetic similarity is equated with close evolutionary relations.
In public health, the emergence of affordable whole genome sequencing of bacterial genomes is currently being developed into a sensing infrastructure for tracking disease. By comparing the genomes of different strains of bacteria epidemiologists and microbiologists now make inferences about how closely related they are. Is this strain of Salmonella close to this other strain? Just as with phylogenetic trees, the logic of genetic disease tracking is that if the genomes are similar, they are seen as related. The logic is that if two organisms are genetically similar, they are thought to share a recent common ancestor. As the leader of the genetics team at the ECDC explained the use of the genetic disease surveillance:
If you have 30 people eating a buffet together and they get sick it is easy to conclude that they belong to the same outbreak. Then you can start to analyse what they have eaten in common. But what do you do in a society where you have maybe 30 000 [cases of] Salmonella per year. What belongs to an outbreak and what does not? (source: personal communication)
In applying the logic of phylogenetics to disease surveillance the genetic methodology is used to link or unlink cases to a specific outbreak.
But before we dive into the enactment of different disease outbreaks, we need small primer on genetics. Genetic disease surveillance is, as outlined above, based on DNA differences between organisms. DNA is said to form the basic genetic blueprints for all living organisms and is also unique for each individual organism. DNA is comprised of four types of molecules, so-called nucleotides, which are paired with each other to form the famed DNA double helix. The DNA helix is comprised of four types of nucleotides: Cytosine, Guanine, Adenine, and Thymine, which are often represented as the letters, C, G, A, and T, when translating the genetic code to letter-form. Thus, DNA strands are often represented as a string of letters: For example, ‘ACGTAA’.
As each individual organism, and in this case each individual Salmonella bacterium, has a unique DNA code, it is possible to identify any individual organism, or bacteria, by analysing its DNA.
A common measure of genetic relation in phylogenetics is to quantify genetic differences by counting differences between different organisms’ DNA. That is, by counting how many nucleotides are different between the DNA of two organisms. A difference of one nucleotide between two organisms is called a Single Nucleotide Polymorphism, a SNP (see image below). This also means that the genetic difference between two organisms, say two Salmonella bacteria, can be quantified by the number of SNPs that set them apart. When one of these nucleotides, one letter in the DNA string, differs between two organisms, that is defined as a one SNP difference.

Fig. 4.2 Two identical DNA strands with one Single Nucleotide Polymorphism, one SNP (source: https://en.wikipedia.org/wiki/File:Dna-SNP.svg)
According to the logic of phylogeny closely related organisms have little genetic difference, while more distant relatives in the genetic tree of life have larger differences. As the head of genetics at the ECDC expresses it:
Few SNPs between different organisms indicate a close relationship and a close common ancestor, a difference of a large number of SNPs indicates a more distant relationship. The number of SNPs that are needed to conclude if the organisms have a close/distant relationship depends on species, type of outbreak, etc. and this is still in the learning phase which can cause interpretation issues within and across sectors.13
The informant thus argued that ‘close’ or ‘distant’ genetic relations between different bacterial strains can be inferred by counting SNP differences. A large number of SNPs is taken as an indication of a close relation, while a small number of SNPs is taken as an indication of distant relation. There is an inference made between genetic similarity and bacterial relation. In essence, the argument is that one can infer that these different bacterial strains are part of the same disease outbreak. In the genetic mode of sensing disease, a disease outbreak is thus enacted by counting SNP differences.
Visualizing genetic similarity at the ECDC
At the ECDC, evidence of disease outbreaks was produced and visualized in varying manners. Geographic intensity maps, and curves of epidemic intensity over time were the most common forms. However, as whole genome sequencing was starting to enter the picture a new type of diagram entered the picture. A visualization of bacterial similarity through detailed phylogenetic trees on the bacterial level. These trees were produced by a genetics team at the ECDC, or in collaboration with expert laboratories in the European Union’s member states. This team was responsible for the collection of sequences generated in member states public health laboratories at the ECDC and collected bacterial isolates from all over the European Union. The team’s goal was to find outbreaks and supporting cross boarder outbreak investigations through genetic evidence.
The most common way of visualizing these genetic linkages at the ECDC was through tree visualizations of genetic relations. In these visualizations every branch on the tree represents a quantified measure of genetic similarity and difference. In the phylogenetic trees of Salmonella, each branching to the right in the figure symbolizes a closer relation between the bacterial strains. In the tree below, this could mean that the rightmost branch might represent a 5 SNP difference, the step to the left, a 10 SNP difference, a step further to the left, a 50 SNP difference, and so on. A vertical line between the bacteria means that the bacteria are identical in terms of genetics, in genetic parlance, they are clonal.
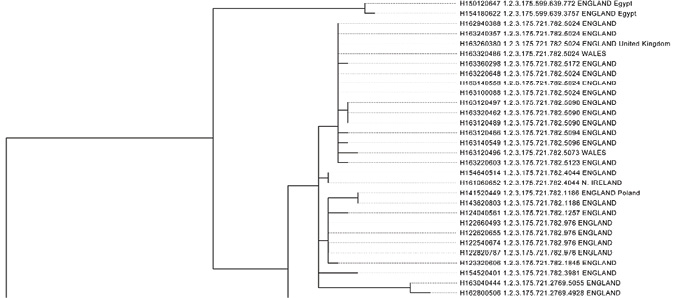
Fig. 4.3 Part of a SNP-based phylogenetic tree of Salmonella Enteritidis (ECDC 2016b: 7)
The phylogenetic trees thus visualize how divergent different bacterial strains were in terms of genetic difference. Each branch on the phylogenetic tree representing a quantified difference of SNPs.
Producing these trees involves assessing and defining the genetic boundaries between one bacterial strain and another, breaking up genetic continuums into quantified and discrete branches of sameness and difference. The production of these trees thus entails translating differences on the genetic level into numerical and graphical representations. This involves deciding how many SNP differences should constitute a new branch on the phylogenetic tree. This also involves assessing what is a big difference and a small difference. How many SNPs is close? And how many SNPs are far away?
Sensing a new class of disease outbreaks
Fig. 4.4 Map of Salmonella cases (ECDC 2019: 3)
One of the consequences of these new genetic groupings of bacteria is that new disease outbreaks become visible to the ECDC. What was before identified as sporadic cases of Salmonella, had now shifted to the identification of regional/national or pan-European outbreaks of Salmonella. By genetically grouping together bacterial isolates sampled in different countries across Europe through the ECDC was starting to see new outbreaks across Europe. What was previously understood as regional outbreaks was now seen as pan European outbreaks.
With the increasing use of whole genome sequencing a new class of outbreaks became visible to the ECDC.
During my fieldwork there was a long-ongoing multi country outbreak of Salmonella in Europe. The outbreak was recurrently brought up for discussion at the daily roundtable meeting as well as in other meetings. In June, the ECDC was cited in FoodQualityNews, as having used Whole Genome Sequencing to identify the outbreak:
The outbreak was detected through WGS [Whole Genome Sequencing] and is characterized by its long duration with relatively low numbers of cases reported intermittently and peaks of re-emergence in late summer/early autumn between 2014 and 2016. In 2017 this pattern changed, with a peak observed in March. (Whitworth 2017)
By using whole genome sequencing to group bacteria into related strains of bacteria, the ECDC and member state’s experts where able to delineate the outbreak and produce a so-called epicurve, a visualization of cases over time, of the outbreak. The genetic grouping of Salmonella made it possible to produce an image of a persistent outbreak of Salmonella in Europe, which had been ongoing for at least three years.
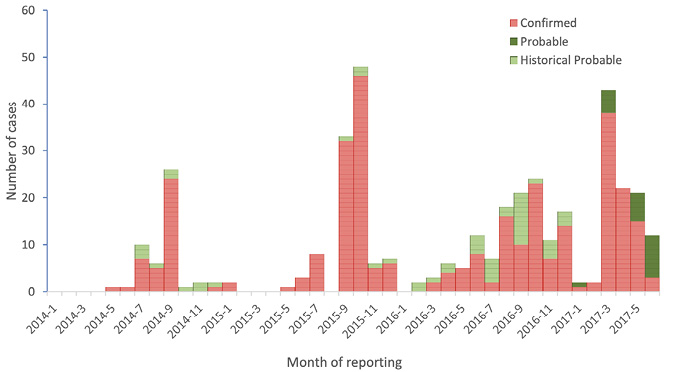
Fig. 4.5 Epicurve of the long ongoing Salmonella outbreak in Country X (ECDC 2017: 4)
The outbreak of Salmonella could now, as my informants phrased it ‘with high precision’, be traced back in time to produce an intensity curve of the outbreak. Thus, genetic evidence was used to produce a previously impossible visualization, an image of how a particular genetic group of Salmonella had spread over Europe. As one of my informants phrased it:
These types of epicurves are classic tools for an epidemiologist. What the new technology provides is much more certainty that the cases are actually true and that the epicurve then represents a true description of the outbreak (given the limitations that are always there in terms of sampling bias/limitations). (Personal communication)
The consequence of whole genome sequencing seemed momentous at the ECDC. The head of surveillance called it a paradigm shift. A whole new set of outbreaks now became visible.
Interpreting genetic sensing infrastructures at the ECDC
However, although the trust in the capabilities of genetic evidence to uncover new outbreaks was strong, the new technology also led to new questions and uncertainties. At the weekly roundtable meeting the team of geneticists recurrently showed phylogenetic trees that genetically grouped bacteria into novel outbreaks. A challenge facing the ECDC at the time of my fieldwork, was that there were no standardized understandings of how bacterial strains mutate, which methods were the most trustworthy, and which nomenclature to describe these things were most fitting.
At the daily roundtable meeting these questions of genetic relation were – for most participants – esoteric questions understood and grappled with by a small team of genetic experts. Consequently, there was a constant struggle to interpret and create meaning from the sometimes obtuse tree visualizations of genetic difference. Both from the experts in the genetics team, but also from disease experts working in different fields. At the daily roundtable meeting – which gathered disease experts with varying degrees of genetic knowledge – questions were constantly posed as a response to the display of these bacterial phylogenetic trees.
At one point, when a representative of the genetics team was scrolling through a seemingly endless phylogenetic tree of bacteria, the chairman of the meeting, who was also the head of disease surveillance, threw up his hands and asked.
Chairman: “But what does it mean?”
Geneticist: “That’s a five SNP difference!”
Chairman: “Is that enough to say it’s the same strain?”
Many different questions of interpreting the phylogenetic trees, and the genetic similarities and differences they represented, were brought forward.
Actor 1: “How fast does Salmonella mutate?”
Actor 2: “Is 5 SNPs a close relation?”
Actor 3: “Is 5 SNPs close enough to declare an outbreak?”
The genetic evidence was not a settled matter for the actors at the ECDC. First, there was an uncertainty about how much genetic difference is a meaningful difference in terms of classifying Salmonella strains. This includes assessing how fast Salmonella mutates, and how stressful the environment is for the bacterium. By settling how stressful the environment is, it was thought that the rate of mutation could be deduced. By settling how fast the bacterial strain mutates, the number of SNP differences could gain meaning. Thus, understanding the bacterium as having a slow rate of mutation implies that only a few SNPs needs to be different for it to be understood as significant difference – and vice versa.
Consequently, there were a number of factors that influenced how genetic likeness was understood. Again, the question that the roundtable was constantly trying to answer was ‘Is this an outbreak of Salmonella?’ Among the complex questions were that different bacteria behave differently, some hardly change over time, while others are very prone to mutation between cases. Another challenge is that if the outbreak is large there is a lot of space for the bacterium to mutate, so you can have wide genetic variance in the same outbreak, which makes sampling technique important, as you can only sample parts of the outbreak ‘branch’ in the phylogenetic tree.
The second question constantly posed at the roundtable meeting, ‘Is 5 SNPs close enough to declare an outbreak?’ is about action and wider consequences of the genetic classification of strains. This is a question of interpreting the data, where the genetics team need to decide if the different bacteria share a ‘recent common ancestor’. Depending on what the team believe is the mutation rate, they can form a hypothesis about two bacteria being part of the same outbreak. The question in their minds is: ‘What is a small enough difference to consider two bacteria as part of the same outbreak?’
The politics of disease surveillance
On the basis of the available evidence, what actions can the ECDC then recommend? Here the work of knowing and constituting an outbreak is shifted and linked to the international politics of food and economies of nations. Just as Gabrys has pointed out, the enactment of particular objects in the world, is closely linked to how people and organizations engage with these objects. The questions at the roundtable meeting must thus be understood against the backdrop of international food politics, where a food-borne disease outbreak can lead to large international repercussions in the form of import bans of different food stuffs. For example, as alluded to above, in 2011, Russia banned the import of cucumbers from the European Union as there was a worry about an outbreak of E. Coli in Germany. Which then also had economic consequences for cucumber farmers. Thus, the economic and political repercussions of disease surveillance were constantly present at the ECDC.
Making the matter even more difficult was the complex organizational situation. The ECDC is in constant collaboration with a number of different organizations around the globe. There is a constant stream of the phone calls, emails, and meetings to coordinate disease surveillance around the world. The ECDC are constantly collaborating with the WHO, the US CDC, and the CDCs of other countries inside and outside the EU. They also collaborate with European organizations within the EU. For example, the European Commission, or the European Food and Safety Authority, EFSA. As Salmonella is a food borne disease, the communication with the European food and safety authority was necessary and legally mandated. This complex organization with different organizational mandates and different political goals makes the introduction of new ways of producing evidence of outbreaks challenging. The question ‘What does a 5 SNP difference mean?’ takes on a whole new pregnancy. The pan-European Salmonella outbreak highlighted all these complex relations between sensing infrastructures, organizations, and new technologies.
The politics of multiple sensing infrastructures
In disease surveillance, theories about the origin of different outbreaks were constantly tested against different sensing infrastructures. For instance, automated algorithms can be pitted against human knowledge and expertise (Lee 2017, 2021), new models of transmission pathways can reshape our understanding of a disease (Lee et al. 2019), or, as in this case, genomic knowledge can be pitted against traditional epidemiological work. As a result, the value of different sensing infrastructures for tracking disease was not apparent at all times at the ECDC. For my informants there was a constant struggle to interpret sensing infrastructures and to determine the source of a disease outbreak: Does the genetic evidence point at a specific country? What did genetic evidence mean in the case of this outbreak?
When modes of sensing add up: The closure of a Polish egg packing facility
A particular outbreak of Salmonella, which was discovered in Europe in 2016, was seen as a landmark for genetic disease surveillance at the ECDC. In this case, genetic evidence was used to identify and shut down one of the largest egg packing factories in Poland. As the ECDC rapid risk assessment phrased it: ‘The available evidence from WGS [whole genome sequencing], food and environmental investigations, as well as from tracing-back investigation of eggs, establishes a link between this multi-country foodborne outbreak and the packing centre B in Poland…’ (ECDC 2016c: 1) The genetic evidence was in harmony with other investigation methods, but it was genetic information that led to the discovery of the outbreak.
In this case the genetic evidence was linked with ‘shoe leather’ epidemiological evidence. As an informant put it during my fieldwork: ‘The genetic information made detection and specific next steps in the investigation possible.’ In this particular case, the genetic sensing infrastructure was in harmony with the older ‘shoe leather’ methods of tracing disease through food networks.
Thinking with Mol’s (2002: 84) conceptual apparatus, which highlights how objects hold together in the face of multiplicities in practice, the investigation of the Polish egg packing facility can be described as if the different sensing infrastructures were coordinated by adding up: there was a coherence between the genetic and ‘shoe leather’ sensing infrastructures in the enactment of the outbreak of Salmonella. As Mol puts it, this ‘form of addition comes with no worries about discrepancies. It does not suggest that tests have a common object. Instead, it takes tests as suggestions for action: one bad test outcome may be a reason to treat; two or three bad test outcomes give more reason to treat.’ (Mol 2002: 84) Thus, in the case of the Polish egg packing facilities the genetic and ‘shoe leather’ sensing infrastructures added up, and there was no need to handle how different objects were enacted differently by different sensing infrastructures.
When modes of sensing don’t add up: Is Country X the source?
However, in connection with the long-ongoing pan-European outbreak of Salmonella which was grappled with during my fieldwork things were not as simple. Here, the different sensing infrastructures did not add up, and the different sensing infrastructures – of genetics and ‘shoe leather’ methods – were pitted against each other in the mores of national and organizational politics. Thus, in this second Salmonella outbreak, there was no identification of a source of the Salmonella outbreak, the different modes of sensing did not add up.
On the one hand, the ECDC team saw the genetic similarity of the different Salmonella strains as strong evidence. In their view the genetic evidence pointed strongly to Country X as the source of the outbreak.14 The genetics team argued that counting SNP differences – that showed close relation between the different cases – was enough evidence to determine the source of the outbreak as being Country X. The ECDC team’s view was that DNA similarity could be used to infer the source of the outbreak. For them, the source of the Salmonella outbreak could be determined based on genetic technology, phylogenetic logic, and correlational thinking.
On the other hand, the European Food and Safety Authority (EFSA) interpreted the strength of the genetic analysis differently. They argued that the evidence produced through the genetic sensing infrastructure was not sufficient for pinpointing the source of the outbreak to Country X. In EFSA’s understanding, tracing foodstuffs to their origins through traditional outbreak investigation methods – ‘shoe leather methods’ – were seen as necessary to determine the source of the outbreak. Thus, in addition to the genetic evidence that ECDC had put forward, EFSA emphasized the need for additional ‘shoe leather’ evidence to ascertain the source of the outbreak. In EFSA’s way of reasoning, only by finding the food pathways of the disease through the global food networks could the source of disease be sufficiently determined.
To EFSA the genetic evidence wasn’t sufficient. Genetic similarity did not imply certainty about the source of disease. The evidence presented by the genetics team was thus not enough to point out Country X as the source of the Salmonella outbreak. Thinking with different modes of sensing, the EFSA argued that a shoe leather mode of sensing was needed in order to substantiate the ECDC’s claims. For my geneticist informants EFSA’s stance led to some frustration. ‘We send people to jail based on genetic evidence’ one of my informants exclaimed frustratedly at one point of the investigation.
In the end, Country X’s government denied that their poultry industry was the source of the outbreak. However, according to my informants, a significant number of chickens were slaughtered after the genetic outbreak investigation had indicated the country as a potential culprit for the Salmonella outbreak. Nevertheless, the source of outbreak that had been detected through the genetic mode of sensing still remained uncertain. The source of the outbreak was not pinpointed and resolved. There was no closure. The two modes of sensing could not be coordinated, and thus created a Salmonella outbreak with an uncertain source. Simultaneous stability and uncertainty. A Salmonella outbreak without a source, a Salmonella outbreak in limbo.
Discussion: Entangling sensing and government action
Disease outbreaks in today’s disease surveillance are enacted through a plethora of different sensing infrastructures. This includes a multitude of technologies and techniques: genetic profiling, satellite imaging, automated image analysis, computer modelling, as well as many algorithms for processing data. The production of risk objects in disease surveillance is thus intimately intertwined with different sensing infrastructures.15
One consequence of this multiplicity is that the emergence of new sensing infrastructures come to enact new patterns of risk and disease. New risk objects come into being on the global stage of disease surveillance. In the case of European disease surveillance, sensing infrastructures enact risk objects on a national, European, and global stage of disease security – implicating both nations and international organizations such as the ECDC, EFSA or the WHO. Another consequence is that, as new sensing infrastructures emerge, old infrastructures keep existing and being used. Thus, new sensing infrastructures, and new enactments of risk objects come to coexist with older ones. Sometimes these enactments coincide, and sometimes they diverge. When they diverge, this can lead to conflicts about which sensing infrastructures, data, and facts about risk objects can be trusted. Should actors trust older more entrenched sensing infrastructures, or should new infrastructures be trusted more?
What is at stake for actors in disease surveillance are questions such as: which sensing infrastructures are trustworthy? What types of sensing infrastructures can be used to identify and trace disease outbreaks in the complex technological, political, and economic arena that disease surveillance operates on? And as a consequence, which risk objects are constructed as real outbreaks, that must be acted on? In the case that I relate above there are two parallel sensing infrastructures vying for organizational trust: on the one hand the traditional sensing infrastructure which draws on traditional ‘shoe leather’ methods and food tracing, and on the other hand the sensing infrastructure that maps the genetic similarity of different bacterial strains. These sensing infrastructures enact outbreaks of Salmonella in different ways, which in our case is also tied to different organizational contexts, mandates and priorities.
The traditional way of tracing food borne disease in disease control has been to trace the origin of food stuffs. As we have seen above, this is done through the mode of sensing that the actors call ‘shoe leather epidemiology’. The focus of these tracing practices is the construction of likely chains of disease transmission which are used to point to an origin of a disease outbreak. By tracing food stuff or food packaging through food distribution networks, actors in disease surveillance construct what they deem to be a likely disease transmission route through the global food distribution network. That is: actors infer the source of the outbreak through its likely route of transportation and transmission. The actors ask: Can we identify the source of the eggshell? And thus of the Salmonella outbreak? Here practices of producing causal chains – creating likely networks of food transmission – are at centre stage.
As we have also seen above, increasingly affordable genetic sequencing technologies have led to the development of an emerging set of sensing infrastructures based on genetic technologies. Many actors in disease surveillance, see this technological development as a new and improved route to detect and handle outbreaks. The focus of these genetic practices is to infer genetic relations of bacterial strains in order to detect outbreaks. In the case above, actors inferred the source of the outbreak through the genetic similarity of different strains of Salmonella. The actors asked: How genetically similar/different are these different bacterial strains? And does this genetic similarity/difference mean that they are closely related?
In dealing with the diverging enactments of the indeterminate and long-ongoing outbreak of Salmonella in Europe, Mol’s vocabulary on the enactment of objects in practice falls short in attempting to describe how multiple sensing infrastructures are handled. In her work, she suggests different manners in which atherosclerosis is maintained as an object in the face of multiplicity in practice: she suggests adding up, distribution, and inclusion to describe how atherosclerosis becomes enacted in hospital practice. But, as I have pointed out elsewhere, just as with many actor-network theory concepts, the focus of her analysis is on the stabilization of facts, technologies, or in this case disease.16 Her focus is on how objects become enacted as real in practice.
However, the long-ongoing Salmonella outbreak which was contested based on different sensing infrastructures was never stabilised. Uncertainty about the source of the outbreak remained. Here, instead of, like Mol, theorizing the enactment, maintenance, or coordination of objects, I have suggested we also need to create a vocabulary for describing conflicts between different sensing infrastructures. Concepts that can be used to describe and analyse the politics of multiple and diverging sensing infrastructures. This would allow a focus on the politics of sensing – and also in this case the politics of sensing disease outbreaks.
Thus, I suggest that we need to create analytical tools that allow for the description and understanding of oppositions, hierarchies, and indeterminacies that arise between sensing infrastructures. This allows us to gain a deeper understanding of objects that do not stabilize. Of objects that remain weak and unstable in the face of multiplicities of sensors and sensing infrastructures.
Here, I propose one such concept – mode of sensing – which would allow a description of certain facets of the politics of sensing infrastructures: Namely how different infrastructures are trusted differently based on different modes of inferring an objects existence. This would allow us to highlight how different sensors are trusted differently in practice. It would also allow analytical purchase on the slippery politics of sensing. My argument is that the two different sensing infrastructures in this case are tied to two modes of sensing disease outbreaks: one which is based on practices of linking and relating – creating what is thought to be causal inferences of disease transmission – and another based on practices of lumping and splitting bacteria into groups – creating phylogenetic trees based on what is said to be genetic correlations.17
Which sensing infrastructures is then trusted, on the basis of which mode of sensing? And in which context? For legal matters to proceed – for example the closing of an egg packing factory – a particular mode of sensing a disease outbreak might be demanded, while in more practical disease prevention work – without legal repercussions – additional modes of sensing might be trusted. Thus, to understand how sensing infrastructures become intertwined with political and organizational contexts, one facet of the puzzle is to understand how different modes of sensing are intertwined with trust and action. Thus, I argue that we need to understand how different modes of sensing become accepted or rejected in complex actor-networks.
Conclusion
To analytically highlight how diverging sensing infrastructures are handled in practice, the chapter has proposed to pay attention to different modes of sensing. This allows moving beyond Gabrys’ observation that different politics take hold with different sensing infrastructures, toward an analysis of the politics of sensing: of how hierarchies, coordinations, divergences, and indeterminacies are handled when sensing infrastructures are used in practice.
The chapter has followed the practices of sensing disease through a genetic sensing infrastructure. In this, it sketched how affordable genetic techniques has led to the development of a novel genetic sensing infrastructure for surveilling disease outbreaks in Europe. It showed how this genetic sensing infrastructure built on a particular mode of sensing disease outbreaks that posited relations between different Salmonella strains through genetic similarity.
The chapter traced how professionals working with disease surveillance at the European Centre for Disease Control and Surveillance used genetics to identify and trace a ‘long-ongoing outbreak of Salmonella’. However, just as Martin and Lynch (2009) have shown elsewhere, the practices of inferring genetic similarity are tied to considerable uncertainties about what genetic similarity really means in practice. Last, the chapter dealt with the politics of multiple sensing infrastructures in disease surveillance. Here the chapter attended to two recent outbreaks of Salmonella in Europe, and what happens when two sensing infrastructures add up, and what happens when they do not.
Thus, the chapter developed an analytical stance which deviates from the age-old construction stories that are told about objects in actor-network theory (see Galis and Lee 2014). In doing this, the chapter set out to pave an analytical path where enactment of objects through sensing infrastructures is not the only possible story to tell. Instead, the chapter analyses how objects remain contested and non-coherent due to divergent sensing infrastructures to find tools, in the words of Haraway (2010), for staying with the trouble.
Sensing infrastructures are not neutral nor innocent. They are implicated in politics of the most crucial kind. They are implicated in questions of international politics, health and illness, life and death. Above we have seen how Salmonella is enacted through different and sometimes contested sensing infrastructures which are embedded in a complex economic, political, and organizational context. An important conclusion one can draw from this chapter is that sensing infrastructures need to be understood in terms of both multiplicity of sensors and non-coherence of the objects that are sensed.
I believe that this approach to analysing sensing infrastructures opens up a road to analysing not only disease surveillance work, but also for analysing sensing infrastructures based on algorithmic calculation as well as Big Data. Which mode of sensing is a particular sensing infrastructure part of? How does this mode of sensing integrate with different contexts of action? As Adrian Mackenzie has aptly asked: how do we swim in this constantly increasing tidal wave of data? (Mackenzie 2014).
The concept of mode of sensing thus, has a potential wide application in understanding how objects are enacted through different sensing infrastructures. Not least in attempting to understand how new infrastructures are integrated or rejected in different settings. By highlighting different modes of sensing, I believe we can understand better how new infrastructures based on for example genomics, satellite imagining, algorithmic processing, computer models, big data or learning machines become integrated in practice. What mode of sensing is a valid measure of a risk object, and in which context?
Acknowledgements
I want to thank the editors (in alphabetical order), Geoffrey Bowker, Nikolaus Poechhacker, and Nina Klimburg-Witjes for insightful comments and criticisms along the way. I also want to thank the anonymous reviewer for insights about doing an infrastructural inversion of Annemarie Mol’s work. The chapter also benefitted from comments by the Cultural Matters Group at Uppsala University led by Tora Holmberg. Additional thanks go to David Moats and Jenny Lee who took the time to read and comment.
Disclaimer
The content of this report does not necessarily reflect the official opinion of the European Centre for Diseases Prevention and Control (ECDC). Responsibility for the information and views expressed in the report lies entirely with the author.