23
Biobank Boxes: Technologies of Population
Susanne Bauer

Fig. 23.1 Freezer tanks at -160°C. Biobank sample storage for a study on nutrition and health in Denmark (photograph: Susanne Bauer, 2008)
Appearance: frosty, cold, opaque, white fog, from mundane to highly secured. Colour: outside: metallic silver with blue lid; inside: white, foggy. Habitat: basements, labs, corridors in research centres or hospitals, specialised facilities. Behaviour: still and frozen when in repository, fluid and mobile when retrieved. Size: diverse, fitting into each other like Russian dolls: small vials (grouped horizontally) in larger boxes, in household-sized freezers or industrial-sized storage tanks and specialised buildings. Migration: immobilised by temperature, embedded in data network; when defrozen, mutable and migrating between labs, circulating and in-forming publics, publications and policies.
Keywords: containing, storing, freezing, defreezing, mobilising, circulating, moving across scales
A mundane basement corridor of an office building, with various ducts on concrete walls. A fireproof door and an alarm system; a security badge is needed; everyone entering the room is documented by time and name of employee and name of visitor. Inside the basement room there are two rows of metallic storage tanks with blue lids. It is quiet except for the low humming of some remote machinery. My guide, a researcher from the biochemistry lab, puts on a long glove before she opens the lid. Nothing is visible inside except the white fog from liquid nitrogen boiling off. She reaches in and brings up an icy metal scaffold – named ‘elevator’ – holding quadratic boxes. The plastic boxes are filled with yet smaller containers – vials holding the frozen sample material, sometimes attached to straws inside, which can be taken out when the material is needed. Each box on the front side of the scaffold and each vial is labelled with an ID number, barcode and project name.
Zooming in on the material organisation of these cold containers reveals ever more boxes in boxes in boxes, like Russian matryoshkas. Boxes make up and organise the materials and methods of research, in this case a large-scale epidemiological study in ten countries: the ‘European Investigation on Nutrition and Cancer’, which has collected specimens and data from more than half a million participants. Citizens recruited for population-based studies gave the blood samples that are now stored in these epidemiological biobanks. The biobank described here belongs to the ‘EPIC’ study of the early 1990s – its acronym was the pitch to launch a comprehensive European resource for research on nutrition and chronic disease, and a research infrastructure for ‘genomic epidemiology’. Since the 1990s genomics has transformed both biomedical and public health research practices and given rise to new research methods in preventive medicine. Biobanks work by reconfiguring and infrastructuring even questions as mundane as whether eating vegetables is good for you. These collections of boxed materials are invested with modernist promises of scientific novelty and hopes for medical cures, as well as fears about all-encompassing biosurveillance and corporate exploitation of personal data. But how do these biobanks work? What other infrastructures and calculative techniques do they combine with? And what will these promissory boxes and the affects that surround them turn into?
My visit to the basement took place in the context of a research project I conducted at Medical Museion, University of Copenhagen. Exploring Copenhagen as a city of biomedicine took me to the National Hospital, to the Danish Cancer Society, to the clinical biochemistry unit at Herlev Hospital, and back to the former Surgeon’s Academy – now the Medical Museion – where my office was located. I was able to sample some of the plastic vials and storage systems used in biobanking, take them back to the museum and juxtapose them with other pieces of medical apparatus. As a curator of recent biomedicine, I became interested in the materiality and artefacts, including the digital ones, of what practitioners referred to as ‘bio-curation’. I was struck by how some samples were stored mundanely in household freezers, while at the same time being re-fashioned as a promissory resource. Since the 1990s, and especially in the Nordic countries, establishing biobank repositories has been part of the busy activity of creating research infrastructures for genomics, loaded with promises of medical progress and future health (Fortun 2008). When zooming in on the practices and infrastructures of biomedicine, old and new, one finds box techniques at their very core. My curatorial interest focused on those infrastructural devices for containment and mobilisation as a point of departure to rethinking the work of ordering that takes place in and with biobanks. How do these specific boxes classify, order, categorise, store away, im/mobilise? And, regarding the banking in biobanks, how do biobank boxings enact, accumulate, manage, and interfere with the value of their content?
Freezing Boxes, Halting Time
Biobanking is about collecting and boxing biological materials by slowing down life processes using temperatures at which metabolism and decomposition come to a halt. The word ‘biobank’ is an umbrella term for a variety of collections; its taxonomies differentiate between kinds of biobanks using categories such as purpose, size, kinds of samples, retrospective or prospective collection, fixed or unlimited period of storage, types of data, security levels, and site of storage (Dove et al. 2012). Different from some of the local biobanks of epidemiological studies, in the UK, Sweden, Norway, and Canada generic ‘national biobanks’ for genomics research have been established in recent decades, and substantial amounts of funding have been put into their logistics and governance. Nature featured the massive endeavour of the UK Biobank bluntly, reporting that ‘in the past few years, more than a half a million people in the United Kingdom have collectively peed into cups, spat into tubes and had needles stuck in their arms’ (Baker 2012). Scientists and research funders alike seem to have been gripped by a collective fever to gather and store DNA and render the population as a frozen repository.
Large-scale funding has been acquired in a promissory rush for anticipated benefits, with spin-offs to develop biobank technologies from freezers to software and recruitment logistics. The need to convince participants and obtain their consent then transforms the project into a logistics task, necessitating strategies to reach the numbers envisaged in the design. Once set up, the repositories continue to enrol funders, health scientists, health policy-makers, and publics. The collective hopes and promises surrounding these intensified box activities seem largely uncontested in Scandinavian societies, where large publics have embraced the quest for intensified biomedical science for better futures and better health. In terms of the modes of knowledge, ‘evidence-based approaches’ using epidemiological techniques all across the research fields of public health, from biomarker studies to health policy evaluations, appealed to scientists and policy-makers alike. The redefinition of modern epidemiology as the ‘study of the occurrence and distribution of health-related states or events in specified populations’ (Porta 2008) resonates with how public health has been reconfigured as managerial ‘outcome research’. Understood as the basic discipline of health research, epidemiological research practices enrol and formalise disease as disease-in-a-population, an entity that can be modelled as an outcome of any set of potential explanatory factors. These risk factors can range from DNA variation to air pollution, from lifestyle to socioeconomic status; multivariate modelling can analyse any variable distilled from inscription devices, testing it as a potential risk factor. These sets of study designs and analytics biometrics hold together epidemiology as a discipline and at the same time are highly mobile. Taken up by biomedicine, they have informed the movement of evidence-based medicine, and reshaped resource allocation for public health, health promotion and health services research.
Epidemiological studies, whether prospective or retrospective, have in common that they work with specific temporal alignments. They work as futuring devices and materialise a mode of ‘living anticipation’, an infrastructural model of preparedness (Adams et al. 2009). Sketching the hopes and resources invested in biobanking for epidemiology, the Nature article (quoted above) concludes that ‘Samples will grow more valuable as data accumulate. In ten years, an estimated 9,000 of the original donors will have developed Alzheimer’s disease, 10,000 will have breast cancer and 28,000 will have died from heart disease’ (Baker 2012: 145). In this way, samples are expected to acquire value with the disease experience of their donators. Technically, so the hopes go, biomarkers of exposure, susceptibility, and disease – the classic triad of epidemiological inquiry – will be identified in those very samples. Epidemiology, now being scaled down into biobanks, has become a molecular endeavour in frozen population boxes, realigned with disease experience data traced over time. How are we to understand these re-visions and extractive practices at molecular levels, and what happens when they are scaled up from molecules to inform healthcare in society?
In an attempt to maximise the scientific value of the repository, the Danish part of the ‘EPIC study’ includes much more than just blood samples; it even stores fat tissue and toenail clippings as material that can later be analysed for chemical pollutants. However, the value usually attributed to the collection resides not in the different material as such, but in its completeness and statistical representativeness. For instance, UK Biobank refrained from collecting tissue biopsies in order to avoid participants dropping out when asked to undergo such invasive procedures. Instead UK Biobank preferred large case numbers; prioritising large numbers over sample variety is in line with the rules of scientific proof that rank statistical validity highest. This sample size optimising and its related trade-offs shapes the biobanks as infrastructures, and in doing so also shapes the kinds of research that they afford in the future. Decisions over sample collecting gamble on method at the expense of closing down and locking in certain options. Moving from vision to actual material collections, biobanks and databases literally infra-structure the conditions of possibility of future knowledge. I argue, however, that since science is constitutively open to change, present hypotheses and analytic techniques may no longer pertain once the envisaged case numbers have been achieved and the frozen samples retrieved. The tension between the stillness of ‘frozen time’ in the collection and the ever incomplete ‘latest apparatus’ of measurement constitutes the speculative economies and dynamics on which scientific endeavours, mediated by boxes, have come to surf.
Things can go wrong in the biobank boxes, too. If samples defreeze uncontrolledly upon blackout or technical failure, the samples are lost. Also, some biomarkers have been shown to be unstable after the freezing and defreezing process (Baker 2012). Even with minor temperature fluctuation, enzyme activity can change biomarker levels. The largest and most high-tech biobanks for long-term storage keep samples at temperatures as low as -196°C and are connected to emergency power systems at the hospital with redundant power back-up. The effort and resources put into establishing national biobanks might indicate that they are seen as being ‘too big to fail’. More recently, in addition to biobank governance standards and ethics protocols, new biobanks have begun to include ‘legacy planning’ as well as inventories of all ‘freeze–thaw events’ (Matzke et al. 2016). These are means of preparedness for any failure that might occur, and they even factor in the potential future need to close down the biobank and redistribute the samples. Medical research, like museums, faces debates over the conditions of collecting and pressure for some mode of restitution of samples, to other repositories or to their owners, especially when related to colonial medicine and anthropology. For future biomedical research, scientists have suggested building on the ideas of ‘open science’ and ‘wiki-governance’ as a model for the regulation of repositories of biological materials, as has already happened in the case of scientific data platforms (Dove et al. 2012). Epidemiologists have long rejected bottom-up and participatory approaches on the grounds of their epistemologies that are committed to ‘ruling out bias’ at all costs. Yet it is worth asking the question what kind of publics are constituted by epidemiological research, and what biobank and epidemiological studies would look like if they followed less top-down principles. Whether and how the change from their risk governance to wiki approaches will shift controversies over evidence and precautions to new publics may depend on more than just rhetoric. Interestingly, many epistemic challenges to these practices seem to come from digital technology itself – the status of which is highly contested within (digital) epidemiology – as big data analytics may modify established techniques of knowledge-making.
Into the Box, Out of the Box
Collecting and storing fluids is laborious, and the flows into and out of boxes differ between biobanks. Some of the large-scale biobanks use robotics technologies, yet there are a lot of minor and older collections with storage in laboratory spaces or freezers on corridors and in basements. The habitats of the freezers (Figure 23.2) are corridors of a clinical biochemistry department in a hospital. Visiting the adjacent busy clinical biochemistry labs offers a glimpse into the cumbersome steps and cycles of collecting, retrieving, and processing samples during the bio-curation process. Before samples enter the freezer box, they undergo a preparation process. They pass centrifuges that separate out different parts of the blood sample, the buffy coat – the DNA-containing part – serum, plasma, and blood cells. Different from the fully automated biobanks that are displayed in industry advertisements like high-tech cathedrals, the storage temperature in these hospital boxes is in the range of household freezers: -20°C or even just +4°C to store DNA before sequencing (Baker 2012). The duration of their storage depends on the research aspirations associated with them. In studies that run over many decades, special advisory boards take the decisions about which purpose justifies actual usage – and wastage – of samples.

Fig. 23.2 Biobank tanks in their habitat (photograph: Susanne Bauer 2008)
The time it may take for frozen samples to move out of containment again, especially in long-term storage units, depends on risk-benefit considerations, protocols, and board decisions. Once it has been decided that samples are to be used, researchers or robots draw up the elevators with samples, identify the labels on the box, and pull out the horizontal plastic boxes and vials or straws holding the specimen. Human data labour combines tracing and linking materials and data, and generating new inscriptions with stored samples. Once they have been released from the logistics of freezers, and having become liquids again, specimens move up level by level and box by box. The unfrozen samples enter the wet lab, where they are channelled into different kinds of measurement apparatus. Some of these are automated – many look like inkjet printers or copy machines, others use ‘gene chips’ as surfaces that work as testing and inscription devices to generate expression signals. The passage through a measurement apparatus, and a series of transformations, reassemble these signals in terms of biological markers of exposure and disease – a new version of the classic epidemiological triad of ‘environment, susceptibility and disease’. This reassembling takes place during the passage of data through various boxes and chains of translation, in which molecular signals become part of a populationalised measure of exposure, susceptibility, or disease – the latter as anticipated events bound to an uncertain future.
To acquire relevance for public health, biomarker concentrations and other inscriptions produced in the wet lab need to be mobilised. How exactly does this take place? It involves another box, this time a calculative box tool that transforms the inscriptions into determinants and potential risk factors, and aligns them with ‘disease-in-the-population’. The box-machine that enacts this mobilisation is the 2x2 table, a quadratic box with four compartments (Figure 23.3). This is a different kind of box – and a rather powerful one. In epidemiological research, this calculative box is the receptacle for numbers; these count numbers of participants in the exposed and non-exposed groups (rows) and diseased versus non-diseased (columns). In this simplest version for binary variables, the contingency table features four boxes that order and denote the number of individuals in a study population. Using this matrix, researchers process the numbers and compute results in terms of aggregate measures of disease and measures of effects, such as relative risks, odds ratios, and absolute risks. Epidemiologists are trained to work with this box; it is a formal tool of quantification and a matrix for linking exposure and disease. The fourfold table also works as leverage to move variables between scales (from the molecular to the social), bringing frozen samples and data in action. The trick of the trade is to transform any situation, issue, or research question into the format of a 2x2 table. Study designs are geared to precisely this; the table provides the calculative technique to lift up the numbers generated by measurement devices including questionnaires. In short, samples are retrieved from storage logistics, channelled through measurement devices, and transformed into inscriptions that then pass through the leverage of the calculative box.

Fig. 23.3 Epidemiological box-apparatus: the 2 x 2 contingency table.
When tracing the material genealogies of calculative devices, it is found that a host of boxes for populations-in-aggregate have populated epidemiological practices. Before biobanks of frozen populations there were different aggregates, including the urn in statistics and the ‘container model’ or, as it has also been called, the ‘bathtub model’ (Figure 23.4). In this textbook image of ‘population reservoirs’, fluid mechanics are evoked to communicate epidemiologic thinking to student audiences. The use of hydraulics as a model is not exclusive to epidemiology; in fact, hydraulic models have been used as teaching devices in economics. The Philipps machine, developed in Britain in the late 1940s, was a hydraulic model used to teach macroeconomics (Morgan and Bouman 2004). In epidemiology, the container model works to promote an understanding of metrics for health in much the same way as the Philipps machine promoted macroeconomics. The model enacts and habituates to the logic of flow and overflow of a liquid population (health) that is amenable to containment. The hydraulic model evokes analogies between epidemiological measures of disease frequency and the physical forces – such as gravity – that act on and in the fluid container. The ‘population aggregate’ is constituted as an object with measurable characteristics – with ‘mortality output’ and ‘birth input’ streams, and specific prevalence and incidence rates, producing a statistical state of health and disease. More than a metaphor, this virtual model conveys a mechanic of population with pertinent characteristics and behaviour – it makes an entity that ‘possesses’ its own rates of disease. The dynamic model of flow and containment lends itself not only to monitoring but also to engineering through prevention measures. Evoking fluid mechanics and a hydraulic model of population (Figure 23.4) enacts the control of input and output as ‘logical choice’. Thinking in terms of a hydraulic model prompts, for instance, technical modification of influx – including metrics such as birth or immigration rates – creating a need for responses to ‘pressures’ and ‘bottlenecks’. These tools infra-structure the population thinking in demography and epidemiology, co-producing scientific modes of thinking that make appear logical or neutral policies that act on, optimise, and control populations. In this sense, the model being used is performative.
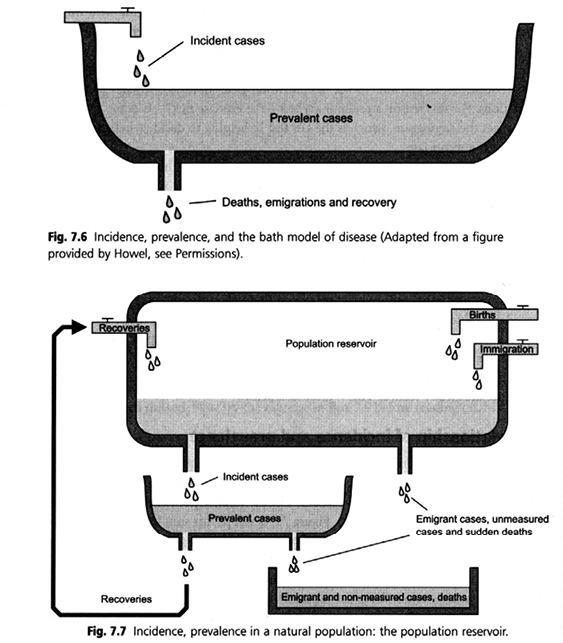
Fig. 23.4 A hydraulic model of population. The ‘container model’ or ‘bathtub model’ (source: Bhopal 2002)
Boxes as Leverage. Containing and Mobilising ‘Population’
Box-tools, like the 2x2 table, work as leverages that mobilise and transpose data into measurable and comparable aggregates. As a consequence, any claim about what is good for health can be channelled into the calculative machine of hypothesis testing that tests the assumption of a causal relation between exposure and disease; this operation results in an estimate of the association between exposure and disease, and a probabilistic test statistic that confirms statistical significance or else attributes the finding to chance. For the use of these calculative devices, it does not matter whether the treated variable is a particular exposure or disease or biomarker, or an economic outcome. These calculative boxes transport methods across domains and scales: no longer confined to risk factor epidemiology, these boxes take part in relating population health to the economic sphere. They enact epidemiology as ‘outcome research’ and as a generalised risk calculation and future-making device. There is hardly any limit to the mobility of these devices; in systematic reviews and meta-analyses epidemiologists even use the very same apparatus to compute an overall risk estimate from the ‘population of results’ from all available studies that meet certain criteria. This population of risk estimate is then formalised in exactly the same way – with the 2x2 table-box – as individuals are treated in an epidemiological study. The double use and scale-mobility of ‘population’ in epidemiology takes off from the scales for which it was developed, adding to and taking in from infrastructures of social sciences and economics.
Following the boxes further upscale takes one to the next box that surrounds the biobank freezers and their contents: the architectural box. That larger box – Herlev hospital – houses clinical staff, patients, researchers, samples, and data generated in clinical trials and epidemiological observation, thereby assembling and infra-structuring people, devices, and knowledge in space and time. Planned in the 1960s, Herlev hospital’s spectacular architecture seems to have much in common with the box principles also found in the biobank (Figures 23.5a, 23.5b).1 The building, designed by architects Gehrdt Bornebusch, Max Brüel and Jørgen Selchau, is known as the highest in the country and referred to as Denmark’s ‘largest piece of art’. Patients’ rooms are located toward the windows, while labs, technical equipment, and services are at the centre and in the adjacent low building. The hospital has been praised, not only for the spectacular view it affords, but also for the interest its architecture holds for patients and staff, among whom stories of ghost floors and hidden tunnels have accompanied praise of its functionality and transparency. This modernist building seems to already belong to the future of a past, but one that can be actualised. In an ‘explosion of colours’ a bright alphanumeric grid is used in its interior as a navigation aid; it is quite striking how these colours correspond with those in the logistics of twenty-first-century test tubes. Both are designed to help navigation and orientation within the abundance of rooms and vials.
In The Birth of the Clinic Foucault describes how new classifications of disease brought about certain spatial organisations and the hospitals of modern medicine (Foucault 1973). So, what are the effects of the biomedical innovations, new numbers, biomarkers, and boxes, and how do they reorganise the clinic? Input and output flows, patient populations, flows of data, capital, and patients are managed side by side with the same calculative devices. Measuring performance indicators, estimating excess, and optimising are part of hospital management, and produce specific biopolitics. Both disciplines measure performance indicators and practise optimising healthcare systems and costs based on these metrics. While epidemiology has been termed the basic science of public health, I argue that logistics has become the basic science of twenty-first-century business operations and organisation. The logistics of scientific knowledge generation include trading and optimising knowledge about risks and benefits against ‘numbers needed to treat’ or ‘numbers needed to harm’ – this is an epistemological necessity that can implicate collateral damage in everyday healthcare settings. The architecture of the late 1960s, which aimed to provide light, colour, openness, hope, and views of a brighter medicine in the hospital, seems like a science-fiction future belonging to an already distant past. While colourful interiors and wayfinding aids convey brightness and clear form, the regimes of economic governance and hospital operation streamline the management of the ‘hospital population’ towards efficiency. Albeit often with different goals in mind, public health researchers and economists evaluate interventions with the same formalisations of aggregates and population. This mode of evaluating in terms of population metrics has reached social policies beyond health, including programmes in social work and poverty reduction. Not last, population reasoning in terms of fluid mechanics often brings about rhetorics of ‘containment’. This is where notions of ‘flood’ and ‘overflow’ gain credibility and are evoked to justify health budget cuts or the closing of borders toward those joining the population. Indeed, techniques of boxing populations in aggregates are susceptible to politics of containment, for instance in health (costs) management in austerity Europe, as well as in recent rhetorics and policies of border enforcement.
Boxes and box technologies shape more than scientific knowledge production; they mediate and mobilize across scale. Biobank practices can lead to different journeys and effects in biomedicine – recombining and the production of novelty, storing away in deep future and falling into oblivion or becoming just waste in a more or less distant future. Upon arrival at the museum, biobank boxes enter yet different kinds of boxes, they might get wrapped in acid free paper or turn into curatorial puzzles about the long-term preservations of plastics. Rather than being package and storage medium as within the biobank, in the curatorial hands they enter the exhibition and then the collection, becoming the object of long-term preservation themselves. And this time it is the boxes and not the samples that are the object of curatorial care – no longer as bio-curating but now redefined as ‘heritage’. In some instances, the boxes themselves might be thrown out as no longer relevant or preserved as infrastructural reminiscence – while becoming heritage they challenge collection and conservation policies. Both types of collections – biobanks and museums – are box-intensive and at times even face similar challenges in terms of changing valuations and issues of accountability, ownership and curatorial care. However, different from the natural history collections in early modern science, their paths seldom cross. It is open what becomes of and what comes after biobanks, how long the pertaining research systems will be active, and when and where they end – whether as waste and ruins of the old infrastructures, aged hypes, oblivion, or recast as heritage in shiny new boxes.
Notes
1 For a presentation of the hospital architecture, see: https://www.herlevhospital.dk/om-hospitalet/Organisation/Sider/Arkitektur-og-udsmykning.aspx [accessed 10 November 2019].
References
Adams, V., M. Murphy, and A. Clarke, ‘Anticipation. Technoscience, Life, Affect, Temporality’, Subjectivity 28 (2009): 246–265.
Baker, M., ‘Building Better Biobanks’, Nature 486 (7 June 2012), 141–46.
Bauer, S., ‘From Administrative Infrastructure to Biomedical Resource: Danish Population Registries, the “Scandinavian Laboratory”, and the “Epidemiologist’s Dream”’, Science in Context 27 (2014): 187–213.
——, M. Fleming, and J. E. Olsén, ‘Im Zwischenraum von Labor und Museum: Eine Ausstellung zur Biomedizin’, in A. te Heesen and M. Vöhringer, eds, Wissenschaft im Museum – Ausstellung im Labor (Berlin: Kulturverlag Kadmos, 2014), pp. 174–95.
Bhopal, R. S., Concepts of Epidemiology (Oxford: Oxford University Press, 2002).
Bowker, G. C., and S. L. Star, Sorting Things Out. Classification and Its Consequences (Cambridge, MA: MIT Press, 1999).
Dove, E. S., Y. Joly, and B. M. Knoppers, ‘Power to the People: A Wiki-Governance Model for Biobanks’, Genome Biology 13 (2012), 158.
Fortun, M., Promising Genomics. Iceland and deCODE Genetics in a World of Speculation (Berkeley, CA: University of California Press, 2008).
Foucault, M., The Birth of the Clinic (New York: Pantheon, 1973).
Matzke, L. A., B. Fombonne, P. H. Watson, and H. M. Moore, ‘Fundamental Considerations for Biobank Legacy Planning’, Biopreservation and Biobanking, 14 (2016), 99–106.
Morgan, M. S., and M. Boumans, ‘Secrets Hidden by Two-Dimensionality: The Economy as a Hydraulic Machine’, in S. de Chadaevian and Nick Hopwood, eds, Models. The Third Dimension of Science (Stanford CA: Stanford University Press, 2004), pp. 369–401.
Porta, M., ed., A Dictionary of Epidemiology, 5th edn (Oxford: Oxford University Press, 2008).
